DNA Extraction from silica gel preserved common bean (Phaseolus vulgaris L.) leaves
Received: March 02, 2018
Accepted: September 01, 2018
Published: November 05, 2018
Genet.Mol.Res. 17(4): http://dx.doi.org/gmr16039926
DOI: http://dx.doi.org/10.4238/gmr16039926
Abstract
Extraction of high-quality DNA from field specimen requires collection under liquid nitrogen which is not readily available in resource constrained laboratories in low- and middle-income countries (LMICs). A method of extracting DNA from silica gel preserved common bean (Phaseolus vulgaris L.) leaves is presented. Our method which does not involve the use of phenol, chloroform or isoamyl alcohol also obviates the need for low temperature incubation during the DNA extraction steps and the grinding of desiccated leaf tissue in liquid nitrogen. It relies on inactivating proteins using SDS and proteinase K and precipitation of polysaccharides using a high salt solution. DNA is further purified by exploiting its insolubility in aqueous media. We were able to extract high quality pure DNA (mean concentration 2.98 ± 0.84 μg/g of leaf tissue) with mean A260/280 of 2.1 ± 0.1 and A260/230 of 2.4 ± 0.15. The DNA was also found to be amenable to amplification using molecular marker types routinely used in molecular biology laboratories like random amplified polymorphic (RAPD) markers, inter simple sequence repeat (ISSR) markers, sequence-characterized amplified region (SCAR) markers and simple sequence repeat (SSR) markers. Our findings show that it is possible to obtain high quality DNA from leaf tissue preserved in silica gel. Our method will be invaluable to resource constrained laboratories especially in LMICs that cannot afford to buy or access liquid nitrogen in order to extract high quality DNA and to research groups undertaking field surveys that require several days or weeks off station without laboratory freezers to maintain the integrity of the tissues which is crucial for obtaining high quality DNA.
Introduction
Good quality deoxyribonucleic acid (DNA) is a pre-requisite for most applications in a molecular biology laboratory. A number of DNA mini prep protocols have been developed and some of them (Agbagwa et al., 2012; Sahu et al., 2012; Aubakirova et al., 2014) rely on modification of earlier sodium dodecylsulphate (SDS) or cetyl-trimethyl ammonium bromide (CTAB) protocols (Rogers and Bendich, 1985; Edwards et al., 1991). Others are fast methods and rely on release of DNA from plant tissue using sodium hydroxide (Wang et al.,1993), by boiling (Thompson and Henry, 1995) or microwave treatment (Saini et al.,1999).
Extraction of DNA from plant tissues using in house DNA extraction buffer formulations generally involves 3 stages; breaking the cell wall to release cellular constituents by grinding tissue in dry ice, liquid nitrogen or fine sand using a pestle and mortar or grinder, disrupting the cell membrane to release DNA into the extraction buffer, use of detergents like SDS or CTAB to solubilize cell wall components and protecting the DNA from nucleases using a chelating like ethylene-diaminetetracetic acid (EDTA). EDTA deprives the nucleases of magnesium ions thereby rendering them inactive.
Separation and denaturation of proteins from DNA is achieved by either emulsifying the buffer- tissue mixture with chloroform and or phenol or by incorporating reducing agents like β-mercaptoethanol, dithiothreitol or sodium sulphite in the DNA extraction buffer. Additional steps to minimize DNA degradation include minimizing turbulence by gently pipetting the DNA solution and minimizing exposure time between pulverization and contact with the DNA extraction buffer. However, despite all these safeguards, producing high quality DNA devoid of degradation remains a big challenge.
To mitigate the effects of degraded DNA, DNA extraction kits have been developed to facilitate the rapid extraction of very highly pure DNA. Such kits use a membrane within a column that binds DNA. Commonly used kits include DNeasy plant mini and maxi kits and the Wizard genomic DNA purification kit and GENEspin (Tamari and Hinkley, 2016). The disadvantage of such kits is that they are not economically feasible for resource constrained laboratories or for laboratories that process a huge volume of samples. In addition, the experimenter has limited options modifying the standard protocols. However, the kits are easy to use, contain all reagents for DNA extraction, require minimal laboratory equipment (usually a centrifuge and micropipettes) and use very few consumables (only pipette tips and micro centrifuge tubes).
Another alternative to the DNA extraction kits is extraction of DNA from paper. A number of proprietary methods of extracting DNA from paper exist and they include IsoCode card (Schelidher and Schuell, Dassel, Germany), generation capture system (Biozym diagnostika GmBH, Hessisch-Oldendorf, Germany), FTA cards (Whatman, Kent, UK). FTA methodology relies on spotting tissue onto the cards and cutting small discs (1.5-2mm) using a tissue biopsy punch of a specific aperture followed by washing of the discs using a proprietary reagent and ethanol. DNA remains impregnated onto the disc which is then used for polymerase chain reaction (PCR) analysis. The method has been successfully used for human forensic studies (Zhong et al., 2001), wildlife DNA (Smith and Burgoyne,2004), PCR based genotyping and plant diagnostics (Drescher and Graner, 2002).
Extraction of high-quality DNA in the laboratory using in-house DNA extraction buffer formulations requires either freshly picked specimen collected under liquid nitrogen for immediate use or specimen collected under liquid nitrogen and stored at -80°C for later use. Both these options are impractical because most field surveys are conducted hundreds of kilometres off station and therefore maintaining the integrity of the tissue to obtain high quality DNA is usually hard. We therefore set out to develop a safe, inexpensive and robust protocol to extract high quality DNA from silica gel dried common bean leaf specimen using equipment that is expected to be available in a basic molecular biology laboratory. We demonstrate the utility of the DNA obtained using common molecular marker systems-simple sequence repeat (SSR) markers, inter simple sequence repeat markers (ISSR), sequence characterized amplified region (SCAR) markers and random amplified polymorphic DNA (RAPD) markers.
Material and Methods
Bean seed was sown into sterilized soils in 5 L plastic pots in the screen house at Kawanda, Wakiso district of Uganda. A trifoliate leaf was picked 10 days after sowing, weighed, wrapped in aluminum foil and put in a sealable plastic bag containing 10 g silica gel. The plastic bag was then put in a Secador desiccator (LabSource, Northlake, IL) and stored for seven days and re-weighed.
Just before DNA extraction, a DNA extraction buffer containing 0.2M Tris-HCl,0.8M NaCl,1% SDS, 20 mM EDTA and 60 μg/mL Proteinase K was prepared. DNA extraction began by grinding the dry leaf in 700 μL of the buffer using a pestle and mortar. The solution was then incubated in a water bath at 65°C for 1h. After incubation, the mixture was centrifuged at 13200 revolutions per min (rpm) for 10 min and 300 μL was transferred to a fresh 1.5 mL tube. Thereafter, 150 μL of 5M NaCl and 900 μL of room temperature absolute ethanol was then added to the supernatant and the mixture was left at room temperature for 2h to precipitate the DNA.
The mixture was then centrifuged at 13200 rpm for 10 min and the supernatant discarded. The resultant pellet was then washed twice with room temperature 70% ethanol which was also discarded. The pellet was then dried at room temperature for 20 min and dissolved in 100 μL of 1X TE buffer (10 mM Tris-HC1 [pH 8], 1 mM EDTA). The DNA solution was then treated with RNase A at a final concentration of 50 μg/mL for 2 h, quantified using the DQ300 fluorometer and immediately used for molecular assays. DNA quality and purity were further assessed by comparing the ratio of the absorbances, A260/280 and A260/230 respectively, using a NanoDrop 2000c spectrophotometer (Thermo Fisher, Wilmington, DE).
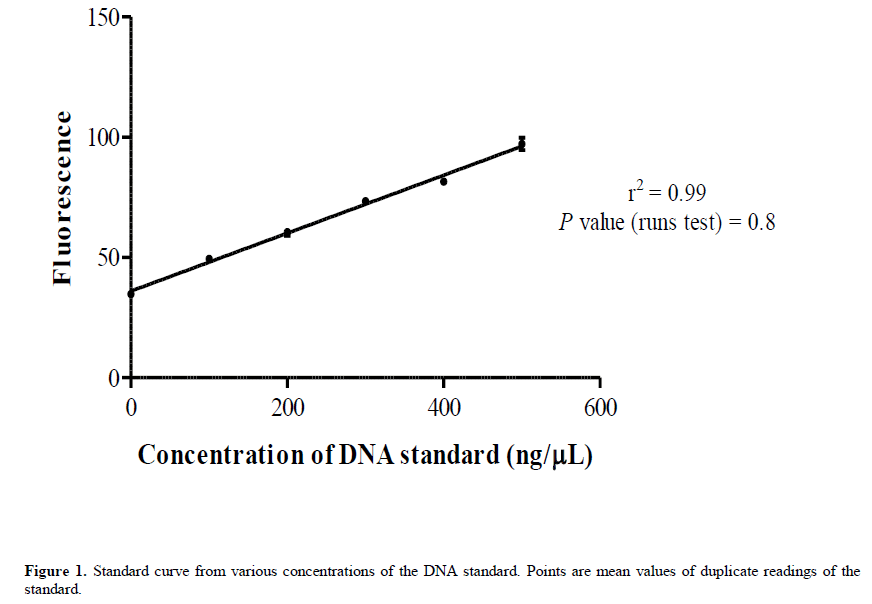
Prior to quantification of the DNA, a standard curve (Figure 1) was generated from various concentrations (100-500 ng/μL) of a known DNA standard (calf thymus DNA) using the DyNAquant DQ 300 fluorometer (Hoefer, Holliston, MA) with the Hoechst 33342 stain (Thermo Fisher, Wilmington, DE) as the fluorescent dye. A Runs test was also performed after the regression analysis to test whether the points significantly differed from linear. Both the standard curve and Runs test were plotted and run using the GraphPad prism software v.5.00 (GraphPad Software, La Jolla California USA, www.graphpad.com).
PCR amplification
All Polymerase chain reactions (PCR) were carried out in 20 μL reactions consisting of 100 ng of DNA, 0.2 mM dNTPs, 2 mM MgCl2, 1X PCR buffer, 0.5U Taq polymerase (Bioneer, Korea) and 0.5 μM of primer (s) For the SSR marker Bmd 502, the PCR reaction mixture was amplified using an initial denaturation at 95°C for 5 and 36 cycles of a touchdown profile each consisting of a denaturation step at 95°C for 20 s, an annealing step at 70°C to 0.5°C each cycle for 20 s and an extension step at 72°C for 30 s. Thereafter a final extension step was performed at 72°C for 5 min and the reaction was stored at 8°C.
The RAPD marker OPV 12 and the ISSR marker RAMS 4 both used a similar thermal cycling profile consisting of an initial denaturation at 95°C for 5 min and 40 cycles each consisting of a denaturation step at 95°C for 20 s, an annealing step at 45°C for 40 s and an extension step at 72°C for 60 s with a final extension step at 72°C for 10 min and storage at 8°C.
The SCAR marker SW13 was amplified with the following thermal profile; initial denaturation at 95°C for 5 min and 30 cycles each consisting of a denaturation at 94°C for 10 s, annealing at 50°C for 20 s and extension at 72°C for 40 s. Thereafter, there was a final extension at 72°C for 5 min and finally, the reaction was stored at 8°C. The marker names, types and sequences used are shown in Table 1.
| Marker Name | Marker sequence (5’- 3’) |
|---|---|
| SW13 | √?¬†F CACAGCGACATTAATTTTCCTTTC R CACAGCGACAGGAGGAGCTTATTA |
| OPV12* | ACCCCCCACT |
| Bmd 502 | F ATTCTCAGGCAGGAAACATA R√?¬† ACGACCCACAATCACTTAAA |
| RAMS 3* | GAG(ACA)5 |
F and R denote forward and reverse primer respectively. Marker systems denoted by * use only 1 primer.
Table 1: Names and sequences of molecular markers used to amplify DNA from silica-gel dried common bean leaves.
Electrophoresis, staining, visualization and gel documentation
The amplicons obtained with the SSR marker were resolved on acrylamide gel using the horizontal polyacrylamide gel electrophoresis (hPAGE) set (Cleaver Scientific, Warwickshire, UK). Briefly, 50 mL of a 6% denaturing polyacrylamide gel consisting of 21 g urea, 1 mL 50X TAE, 7.5 mL of 40% acrylamide-bis acrylamide mix, 100 μL TEMED and 500 μL of 10% APS was made. The solution was then cast onto a hPAGE gel casting unit and a quarter plate overlaid. The mixture was left at room temperature for 30 min to allow complete polymerization of the acrylamide gel mixture. Electrophoresis was performed at 100 V in 1X TAE (0.04 M Tris – Acetate, 0.0001 M EDTA) for 2 h after which the gel was put to stain in 0.5 μg/mL ethidium bromides for 40 min. Finally, the image was captured using the G: BOX gel documentation system (Syngene, Fredrick, MD).
For the ISSR, RAPD and SCAR markers, amplicons were resolved on 1.5% agarose gel in 1X TAE at 100 V for 1 h. The staining procedures and image documentation were like for the SSR marker.
Results
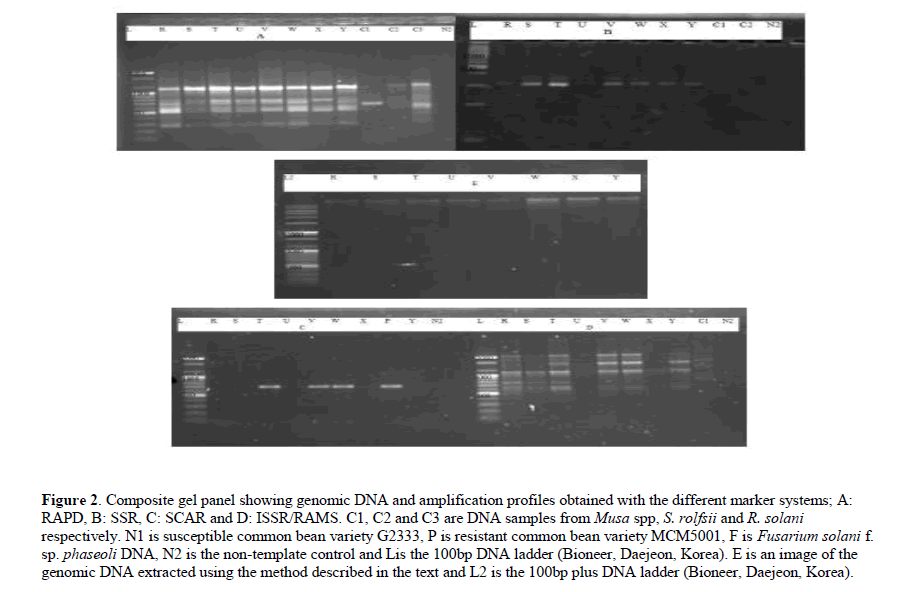
Clear, non-viscous DNA solution was obtained after dissolving the DNA pellet in the TE buffer. In addition, the DNA showed minimum degradation and it was amplified by all the molecular markers assayed (Figure 2)
Figure 2: Composite gel panel showing genomic DNA and amplification profiles obtained with the different marker systems; A: RAPD, B: SSR, C: SCAR and D: ISSR/RAMS. C1, C2 and C3 are DNA samples from Musa spp, S. rolfsii and R. solani respectively. N1 is susceptible common bean variety G2333, P is resistant common bean variety MCM5001, F is Fusarium solani f. sp. phaseoli DNA, N2 is the non-template control and Lis the 100bp DNA ladder (Bioneer, Daejeon, Korea). E is an image of the genomic DNA extracted using the method described in the text and L2 is the 100bp plus DNA ladder (Bioneer, Daejeon, Korea).
The readings for the DNA concentration were very accurate since they were derived from the standard curve (Figure 1) which showed similar accuracy. The standard curve had a high value of r2 (0.99) and a nonsignificant p-value (0.8) of the Runs test indicated the suitability of linear regression in fitting the curve.
The mass of the fresh leaves ranged from 1.97-2.27 g (mean=2.09 ± 0.098), the mass of the dried leaves ranged from 1.91-2.12 g (mean=2.01 ± 0.078) and the percentage change in mass ranged from 2-7.7% (mean =0.04 ± 0.02). These values are shown in Table 2.
| Sample ID | Fresh mass (g) | Dry mass (g) | %√?¬† change in mass |
|---|---|---|---|
| X | 2.27 | 2.12 | 0.066 |
| U | 1.98 | 1.94 | 0.020 |
| R | 2.13 | 2.04 | 0.042 |
| V | 2.07 | 2.02 | 0.024 |
| S | 2.12 | 2.08 | 0.019 |
| Y | 2.15 | 2.06 | 0.042 |
| T | 2.08 | 1.92 | 0.077 |
| W | 1.97 | 1.91 | 0.030 |
| Mean | 2.09(0.098) | 2.01(0.078) | 0.04(0.02) |
Values in parentheses are standard error of the mean of the observations
Table 2: Mass and percentage change in mass observed among eight common bean lines.
Overall, the total amount of DNA per gram of dry tissue ranged from 1.92-4.5 μg (mean=2.98 ± 0.84). The measure for purity of DNA indicated by the ratio of the absorbance at 260 nm to absorbance 280 nm (A260/280) ranged from 1.98-2.31 (mean=2.1 ± 0.1) while the ratio of the absorbance at 260 nm to absorbance 230 nm (A260/230) that indicates protein concentration ranged from 2.13-2.54 (mean =2.4 ± 0.15). The values associated with DNA are shown in Table 3.
| ID/Mean | DNA (√?¬Ķg/g) | A260/280 | A260/230 |
|---|---|---|---|
| X | 2.24 | 2.31 | 2.13 |
| U | 2.88 | 2.09 | 2.49 |
| R | 3.09 | 2.13 | 2.16 |
| V | 4.51 | 1.98 | 2.34 |
| S | 2.56 | 2.19 | 2.50 |
| Y | 3.83 | 2.10 | 2.54 |
| T | 1.91 | 2.03 | 2.34 |
| W | 2.85 | 2.07 | 2.39 |
| Mean | 2.98(0.84) | 2.1(0.1) | 2.4(0.15) |
DNA concentration was determined by fluoroscence using the Hoechst dye.√?¬† Values in parentheses are standard error of the mean of the observations
Table 3: Average tail length and standard deviation (√?¬Ķm) in SCAMCN of M√?¬©xico and South America (86 accessions from Mexico and 27 from South America) exposed to two different concentrations of nicosulfuron (40,000 and 80,000 ppm) and the negative controls.
Discussion
The objective of this study was to develop a DNA extraction protocol for the recovery of high-quality DNA from field specimen. A moderate amount of DNA was extracted. Our findings are lower than those of Agbagwa et al. (2012) who obtained up to 10 μg of DNA per gram of tissue of cajanus species. The reason for the difference in the amount of DNA recovered could be the method of quantification. The study by Agbagwa et al. (2012) used spectrophotometric methods to quantify the DNA while our study used fluorescence. Spectrophotometric (absorbance) methods overestimate the amount of DNA by quantifying single stranded DNA and RNA alongside double stranded (ds) DNA (O’Neil et al., 2011; Sironen et al.,2011) but fluorescent dyes bind only to dsDNA.
The DNA solution was clear and not viscous. These observations are indicative of the quality of the DNA and they are supported by our values for the measures for DNA purity and absence of contamination by proteins being within the ranges stipulated for highly pure DNA (A260/230 =1.7-2, A260/230 >2 respectively (Zeugin and Hartley, 1985; Crouse and Amorese, 1987). The clarity of the DNA is suggestive of absence of contaminating secondary metabolites especially polyphenolic compounds. These compounds impart a brown-red color to the DNA pellet resulting in a solution of similar color (Katterman and Shattuk, 1983). A non-viscous DNA solution is indicative of minimum contamination of DNA with polysaccharides (Do and Adams, 1991). We advance two reasons for the low levels of contaminants. The age of the tissue used and use of high concentration of NaCl in the extraction buffer and in the precipitation of DNA (instead of sodium, potassium or ammonium acetate). Young leaf tissues contain low levels of secondary metabolites like polysaccharides and polyphenols while NaCl not only minimizes the formation of polysaccharides but also prevents them from co-precipitating with the DNA (Fang et al., 1992).
The DNA obtained showed minimal degradation. We believe this was due to activity of the DNases during the maceration with sand. Labuza (1970) reviewed properties of water under food preservation conditions and concluded that the water in food does not leak out unless there is damage to the membranes. Basing on this observation we want to think that drying under silica gel is somewhat stringent resulting in the lysis of the membranes. DNases then gain access to the DNA causing its degradation. Macerating the tissue in liquid nitrogen can reduce degradation.
Conclusion
We have demonstrated an inexpensive, reliable and scalable method that recovers good quality DNA from field specimen without prior preservation in liquid nitrogen. Our method will find particular utility in resource constrained laboratories in low- and middle-income countries (LMICs) that are keen on using marker assisted selection (MAS) in their breeding activities.
Acknowledgments
The authors would like to thank CIAT for funding this study. The technical support provided by the CIAT staff (Musoke Stephen, Hasifa Nababi and Samuel Kiviri) is also highly appreciated.
About the Authors
References
- Ahmadi J, Mohammadi A and Mirak N (2012). Targeting promising bread wheat (Triticum aestivum L.) lines for cold climate growing environments using AMMI and SREG GGE Biplot analyses. J. Agric. Sci. Technol. 14: 645-657.
- Allard RW (1971).Princ√?¬≠pios do Melhoramento Gen√?¬©tico das Plantas. USAID/Edgard Blucher, Rio de Janeiro.
- Alvarez MP and Eyh√?¬©rabide GH (1996). Estabilidad del rendimento de cultivares de h√?¬≠bidoscomerciales de ma√?¬≠z em la √?¬°rea de la EEA Pergamino. Rev. Tecnol. Agropec 5: 17-21.
- Amorim EP, Camargo CEO, Filho AWPF, Junior AP, et al. (2006). Adaptabilidade e estabilidade de linhagens de trigo no Estado de S√?¬£o Paulo. Bragantia 65: 575-582 https://doi.org/10.1590/S0006-87052006000400007.
- Annicchiarico P (1992). Cultivar adaptation and recommendation from alfafa trials in Northern Italy. J. Gen. Plant Breed 46: 269-278.
- Benin G, Pinnow C, Silva CL, Pagliosa ES, et al. (2012). An√?¬°lises biplot na avalia√?¬ß√?¬£o de cultivares de trigo em diferentes n√?¬≠veis de manejo. Bragantia 71: 28-36 https://doi.org/10.1590/S0006-87052012000100005.
- Campbell BT and Jones MA (2005). Assesment of genotype x environment interactions for yield and fiber quality in cotton performance trials. Euphytica 144: 69-78. https://doi.org/10.1007/s10681-005-4336-7
- Castillo D, Matus I, Pozzo A, Madariaga R, et al. (2012). Adaptability and genotype times environment interaction of spring wheat cultivars in Chile using regression analysis, AMMI, and SREG. Chil. J. Agric. Res. 72: 167. https://doi. org/10.4067/S0718-58392012000200001
- CONAB - Companhia Nacional de Abastecimento (2012). Acompanhamento da safra brasileira de gr√?¬£os. 3√?¬į Levantamento Gr√?¬£os Safra 2012/13 - Setembro 2012. Available at [http://www.conab.gov.br/OlalaCMS/uploads/ arquivos/14_01_10_10__12_36_boletim_portugues_dezembro_2012.pdf]. Accessed October 7, 2016.
- Cond√?¬© ABT, Coelho MAO, Yamanaka CH and Corte HR (2010). Adaptabilidade e estabilidade de gen√?¬≥tipos de trigo sob cultivo de sequeiro em Minas Gerais. Pesqui. Agropecu. Trop. 40: 45-52.
- Cruz CD (2013). GENES - a software package for analysis in experimental statistics and quantitative genetics. Acta Scientiarum 35: 271-276.
- Cruz CD, Carneiro PCS and Regazzi AJ (2014). Modelos biom√?¬©tricos aplicados ao melhoramento gen√?¬©tico. 3 ed. Editora da UFV, Vi√?¬ßosa.
- Da Silva GO, De Carvalho ADF, Souza ZS, Ponijaleki RS, et al. (2016). Desempenho genot√?¬≠pico de clones de batata via modelos mistos. Agraria 11: 259-266 https://doi.org/10.5039/agraria.v11i4a5390.
- Duarte JB and Vencovsky R (1999). Intera√?¬ß√?¬£o gen√?¬≥tipo x ambiente: uma introdu√?¬ß√?¬£o √?¬† an√?¬°lise “AMMI”. Sociedade Brasileira de Gen√?¬©tica, Ribeir√?¬£o Preto.
- Junior OP (2014). Aptid√?¬£o, adaptabilidade e estabilidade fenot√?¬≠pica de gen√?¬≥tipos de trigo. Disserta√?¬ß√?¬£o (Mestrado em Agronomia) - Departamento de Produ√?¬ß√?¬£o Vegetal da Universidade Estadual do Centro-Oeste (UNICENTRO), Guarapuava.
- Ma BL, Yan W, Dwyer LM, Fregeau-Reid J, et al. (2004). Graphic analysis of genotype, environment, nitrogen fertilizer, and their interactions on spring wheat yield. Agron. J. 96: 169-180.
- Oliveira AB, Duarte JB and Pinheiro JB (2003). Emprego da an√?¬°lise AMMI na avalia√?¬ß√?¬£o da estabilidade produtiva em soja. Pesqui. Agropecu. Bras. 38: 357-364 https://doi.org/10.1590/S0100-204X2003000300004.
- Oliveira DM, Souza MA, Rocha VS, Assis JC, et al. (2011). Desempenho de genitores e popula√?¬ß√?¬Ķes segregantes de trigo sob estresse de calor. Bragantia 70: 25-32 https://doi.org/10.1590/S0006-87052011000100005.
- Pereira HS, Melo LC, Peloso MJD, Faria LC, et al. (2009). Compara√?¬ß√?¬£o de m√?¬©todos de an√?¬°lise de adaptabilidade e estabilidade fenot√?¬≠pica em feijoeiro-comum. Pesqui. Agropecu. Bras. 44: 374-383 https://doi.org/10.1590/S0100-204X2009000400007.
- Pereira TCV, Schmit R, Haveroth EJ, Melo RC, et al. (2015). Reflexo da intera√?¬ß√?¬£o gen√?¬≥tipo x ambiente sobre o melhoramento gen√?¬©tico de feij√?¬£o. Cienc. Rural 46: 411-417 https://doi.org/10.1590/0103-8478cr20130998.
- Piepho HP, Williams ER and Maddlen LV (2012). The use of two-way linear mixed models in multitreatment meta-analysis. Biometrics 68: 1269-1277.
- Pimentel Gomes F (2009). Curso de estat√?¬≠stica experimental. 15 ed. FEALQ, Piracicaba.
- Pupin S, Dos Santos AVDA, Zaruma DUG, Miranda AC, et al. (2015). Produtividade, estabilidade e adaptabilidade em prog√?¬™nies de poliniza√?¬ß√?¬£o aberta de Eucalyptus urophylla ST Blake. Sci. For. 43: 127-134.
- R Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- Ramalho MAP and Ara√?¬ļjo LCA (2011). Breeding self-pollinated plants. Crop Breed. Appl. Biotechnol. 11: 1-7 https://doi.org/10.1590/S1984-70332011000500002.
- Ramalho MAP, Ferreira DF and Oliveira AC (2000). Experimenta√?¬ß√?¬£o em gen√?¬©tica e melhoramento de plantas. UFLA, Lavras.
- Resende MDV (2002). Gen√?¬©tica biom√?¬©trica e estat√?¬≠stica no melhoramento de plantas perenes. Embrapa Informa√?¬ß√?¬£o Tecnol√?¬≥gica, Bras√?¬≠lia. Embrapa Florestas, Colombo.
- Resende MDV (2007). Software SELEGEN-REML/BLUP: sistema estat√?¬≠stico e sele√?¬ß√?¬£o gen√?¬©tica computadorizada via modelos lineares mistos. Embrapa Florestas, Colombo.
- Resende MDV (2016). Software Selegen-REML/BLUP: a useful tool for plant breeding. Crop Breed. Appl. Biotechnol. 330-339 https://doi.org/10.1590/1984-70332016v16n4a49.
- Resende MDV and Duarte JB (2007). Precis√?¬£o e controle de qualidade em experimentos de avalia√?¬ß√?¬£o de cultivares. Pesqui. Agropecu. Trop. 37: 182-194.
- Rodr√?¬≠guez GRE, Medina PJ, Puente EO, Reyes LA, et al. (2011). Interacci√?¬≥n genotipo-ambiente para la estabilidad de rendimiento en trigo en la regi√?¬≥n de Mexicali, BC. M√?¬©xico. Trop. Subtrop. Agroecosyt 14: 543-558.
- Rosado AM, Rosado TB, Alves AA, Laviola BG, et al. (2012). Sele√?¬ß√?¬£o simult√?¬Ęnea de clones de eucalipto de acordo com produtividade, estabilidade e adaptabilidade. Pesqui. Agropecu. Bras. 47: 964-971 https://doi.org/10.1590/S0100-204X2012000700013.
- Silva RR and Benin G (2012). An√?¬°lises Biplot: conceitos, interpreta√?¬ß√?¬Ķes e aplica√?¬ß√?¬Ķes. Cienc. Rural 42: 1404-1412 https:// doi.org/10.1590/S0103-84782012000800012.
- Silva RR, Benin G, Silva GO, Marchioro VS, et al. (2011). Adaptabilidade e estabilidade de cultivares de trigo em diferentes √?¬©pocas de semeadura, no Paran√?¬°. Pesqui. Agropecu. Bras. 46: 1439-1447 https://doi.org/10.1590/S0100-204X2011001100004.
- Silveira DA, Princinotto LF, Nardino M, Bahry CA, et al. (2016). Determination of the adaptability and stability of soybean cultivars in different locations and at different sowing times in Paran√?¬° state using the AMMI and Eberhart and Russel methods. Semin. Cienc. Agrar. 37: 3973-3982 https://doi.org/10.5433/1679-0359.2016v37n6p3973.
- Spilke J, Phiepho HP and Hu X (2005). Analysis of unbalanced data by mixed linear models using the Mixed Procedure of the SAS System. J. Agron. Crop Sci. 191: 47-54.
- Streck EV, K√?¬§mpf N, Dalmolin RSD, Klamt E, et al. (2008). Solos do Rio Grande do Sul. 2 ed. Emater, Porto Alegre. Yan W and Kang MS (2003). GGE biplot analysis: A graphical tool for breeders, geneticists, and agronomists. CRC Press, Boca Raton.
- Yokomizo GKI, Duarte JB, Vello NA and Unfried JR (2013). An√?¬°lise AMMI da produtividade de gr√?¬£os em linhagens de soja selecionadas para resist√?¬™ncia √?¬† ferrugem asi√?¬°tica. Pesqui. Agropecu. Bras. 48: 1372-1380 https://doi.org/10.1590/ S0100-204X2013001000009.
- Zobel RW, Wright MJ and Gauch HG (1988). Statistical analysis of a yield trial. Agron. J. 80: 388-393.
Keywords:
Download:
Full PDF- Share This