Differential expression of genes associated with low-level vancomycin resistance in Staphylococcus aureus
Received: May 01, 2019
Accepted: May 06, 2019
Published: July 05, 2019
Genet.Mol.Res. 18(2):
Keywords
Antimicrobial susceptibility; Staphylococcus aureus; Vancomycin resistance; Relative gene expression; Heterogeneous vancomycin-intermediate S. aureus
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an important cause of serious bacterial infections. The widely used of vancomycin for many decades leading to the immerging of MRSA with intermediate resistance to vancomycin (VISA) in 1997 (Hiramatsu et al., 1997). Though such strains are designated as ‘intermediately resistant’, they are practically resistant to the concentrations of vancomycin used in clinical therapy (Fridkin et al., 2003). VISA strains are frequently generated from MRSA strains which are heterogeneously resistant to vancomycin or hetero-VISA (hVISA). The hVISA isolates contain sub-populations that exhibit reduced susceptibility to vancomycin while their vancomycin minimum inhibitory concentrations (MICs) are within the susceptible range (<2 μg/mL) (Hiramatsu, 2001). These organisms are clinically important because they can persist in hospital environments and become VISA during vancomycin therapy in patients infected with hVISA (Liu, 2003). Now-a-days, hVISA cannot be detected by routine susceptibility test, thus several screening methods have been proposed (Satola et al., 2011). A confirmatory test of population analysis profile with area under the curve (PAP-AUC) is still needed albeit it is not appropriate for routine service because it is laborious and time-consuming (Wootton et al., 2001; Howden et al., 2010). The resistance mechanism of the hVISA and VISA phenotypes is related to the functional alteration of several metabolic genes especially those involving the alteration of cell wall biosynthesis and thickening (Cui et al., 2003). The specific genes contributing to low-level vancomycin-resistance have not yet been elucidated. Cumulative mutations in diverse genetic pathways have been found when vancomycin-susceptible strains were transformed to hVISA and VISA (Mwangi et al., 2007; Howden et al., 2008; Cui et al., 2009). Several point mutations in the regulatory system modulating cell wall metabolisms were reported (i.e., the walRK, graSR and agr quorum sensing system) (Mwangi et al., 2007; Howden et al., 2008; Cui et al., 2009). Proteomic analysis of hVISA demonstrated that there is an up-regulation of isaA gene compared to vancomycin-susceptible S. aureus (VSSA) (Stapleton et al., 2007; Matsuo et al., 2011; Chen et al., 2013). In the current study, the respective quantitative expression of 5 genes (isaA, atl, walK, walR, pbp4) related to bacterial cell wall metabolism and one related to peptide formation (rplN) were determined among hVISA, VISA and VSSA isolates. This study could provide a broad view of the expression level of these selected genes among S. aureus strains with different resistance to vancomycin and might be useful in categorization of hVISA, VISA and VSSA.
Materials and Methods
Ethics approval
This study was approved by the Ethics Committee of Khon Kaen University (HE552272).
Bacterial strains
A total of 69 MRSA isolates collected from patients at Srinagarind Hospital between 2002 and 2011 were studied. All isolates were identified by Gram staining and biochemical testing (catalase, coagulase, DNase, and mannitol fermentation) and confirmed by detection of either femA or nuc gene using PCR (Berger-Bachi et al., 1989). Methicillin-resistance was identified by detection of the mecA gene using PCR (Kondo et al., 2007). The strains were determined for Staphylococcal Cassette Chromosome (SCC) mec types and agr types according to previous reports (Lina et al., 2003; Kondo et al., 2007). Genomic DNA used for all amplifications was extracted using achromopeptidase (Wako Chemicals, USA) (Shittu et al., 2004). Each isolate was classified as VSSA, hVISA and VISA by determining the minimum inhibitory concentration (MIC) of vancomycin by using an agar dilution method, together with one point analysis, a screening method for hVISA and VISA, and then confirmed by PAP-AUC (Wootton et al., 2001) using Graph Pad Prism 5.0.1 (GraphPad Software Inc., San Diego, USA) for determining the AUC. The PAP-AUC ratios of the test isolate and that of the reference hVISA isolate (Mu3) were calculated. The isolate which had PAP-AUC ratio of 0.90 –1.30 and > 1.30 was interpreted as hVISA and VISA respectively. All isolates were kept at -20°C in skimmed milk containing 20% glycerol until used. S. aureus ATCC29213, S. aureus ATCC700698 (Mu3), and S. aureus ATCC700699 (Mu50) were used as the control VSSA, hVISA and VISA strains, respectively.
Antimicrobial susceptibility testing
The MICs to vancomycin for all of the isolates was determined by an agar dilution method according to the CLSI standard procedure (Clinical and Laboratory Standards Institute, 2010).
RNA extraction and cDNA synthesis
An aliquot of overnight culture was diluted 1:50 in Trypic soy broth (Oxoid, UK) and grown to the exponential phase, which was determined by measuring the OD600 at 0.4 (The Ultrospec™ 1100 pro UV/visible spectrophotometer, GE Healthcare). The bacterial cells were harvested and lysed by incubation with 10 μLof 10 U achromopeptidase at 55°C for 10 min. After incubation, the total bacterial RNA was extracted from the other bacterial components using TRIzol® Reagent (Invitrogen, Australia). The bacterial RNA was re-suspended in 30 μL RNase-freewater (Invitrogen). The contaminated bacterial DNA was further eliminated using the Turbo DNA-free Kit (Invitrogen) as per the manufacturer’s instructions.
The cDNA was synthesized using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystem). Ten microliters of total RNA was reverse transcribed according to the manufacturer’s instructions in a total volume of 20 μL. All cDNA samples were stored at -20°C until used.
Quantitative Real-Time PCR
Relative gene quantifications were performed using a QuanStudioTM 6 Flex Real-Time PCR System (Applied Biosystems). Primers were designed from the corresponding gene sequences derived from GenBank sequences (http://www.ncbi.nlm.nih.gov) using Beacon Designer 8.12 software. The multiple sequence alignment was done using Florence Corpet (http://multalin.toulouse.inra.fr/multalin/). The OligoAnalyzer 3.1 (http://sg.idtdna.com/calc/analyzer) was subsequently used to analyze for potential hairpin-loops, primer-dimers, and hetero-dimers. Gene specificity of all primers was confirmed using BLAST (http://ncbi.nlm.nih.gov/BLAST). All sequences of the primers used in the current study are shown in Table 1.
| Gene | Forward primer (5’ to 3’) | Reverse primer (5’ to 3’) | Locationa |
|---|---|---|---|
| isaA | GCAGTATTACCATTGCTTA | GCACAATCAAGTAACTCA | 2648126-2648827 |
| atl | ACACCGTTTAATGATGAAA | GTTGGGTAAGTAAAGCATA | 1026342-1030088 |
| rplN | GTGCTAATGATACGATTTTCA | TGACGGTTCATACATCAA | 2303125-2303493 |
| pbp4 | CGGCATTACTAGAATTAGATA | CTACCTGAGTTGAGTAATAC | 690688-691983 |
| walK | GTGACTGAACAACAACAA | TGGTGCTAGTTCTTCATC | 25100-26926 |
| walR | CCAGACATCGTATTATTAGATA | GCAGTAAGCATTATAATTGG | 24386-25087 |
| 16S rRNA | GCCTAATACATGCAAGTC | CCAGTCTTATAGGTAGGTTA |
a. Sequences are derived from GenBank accession number BA000018.3 for the pbp4, atl, rplN, isaA, and 16S rRNA genes and CP002114.2 for the walk and walR genes. The location refers to the position of each amplicon located within its corresponding sequence.
Table 1: List of primers for real-time quantitative reverse transcriptase – PCR
Real-time PCR reaction was performed using Power SYBR® Green PCR Master Mix (Applied Biosystem). The total volume of each reaction was 20 μL, containing 10 μL of Power SYBR® Green PCR Master Mix, 2 μL of cDNA template, and 0.2 μM final concentration of each primer. The cycle conditions were: 10 min at 95°C; followed by 45 cycles of 20 s at 95°C; 20 s at 53°C; and 30 s at 72°C and a final cycle of 15 s at 95°C; 1 min at 60°C; and 15 s at 95°C. The expression experiment was done in triplicate. The relative quantification (RQ) of gene expression was calculated based on the comparative Ct method (Livak and Schmittgen, 2001) using Qunastudio Real-Time PCR Software version 10.2 (Applied Biosystem®). In brief, the mean of the replicated Ct values of each tested genes were normalized by subtraction with those of their 16S rRNA gene acted as internal control gene. These normalized values were then subtracted to those normalized values of the respective gene of the control VSSA strain (i.e., S. aureus ATCC29213) and displayed as the RQ value.
Statistical analysis
All data are presented as means ± their respective standard error of the mean (SEM). Statistical analyses were performed using One-Way ANOVA followed by an least significant difference (LSD) post-hoc test using SPSS version 19.0. A p value of <0.05 was considered statistically significant.
Results
Characteristics of the bacterial isolates
All 69 MRSA isolates were found to carry the femA and mecA genes. The respective PAP-AUC ratio ranges for the VSSA, hVISA and VISA groups were 0.42-0.87, 0.9-1.27, and 1.35-3.91. These represented 26 VSSA, 28 hVISA and 15 VISA isolates. The respective ranges for vancomycin MICs were between 1-2, 1-3, and 4- >16 μg/mL (Table 2). The genotypes of the isolates were SCCmec III agr I for 49 isolates, SCCmec II agr II for 15 isolates, SCCmec II agr I for 3 isolates and SCCmec III agr II for 2 isolates.
| Isolates (n) |
PAP-AUC ratio range (mean ± SEM) |
Vancomycin MIC range (MIC50/90 ) µg/ml |
|||||
|---|---|---|---|---|---|---|---|
| VSSA (26) | 0.42-0.87 (0.61 ± 0.03) | 1-2 (1/2) 1-3 (1/2) 4->16 (7/15) |
|||||
| hVISA (28) | 0.9-1.27 (0.99 ± 0.02) | ||||||
| VISA (15) | 1.35-3.91 (2.26 ± 0.30) | ||||||
Table 2: Phenotypic characteristics of the VSSA, hVISA, and VISA isolates
Relative quantification of the 6 genes in the VISA, hVISA, and VSSA groups
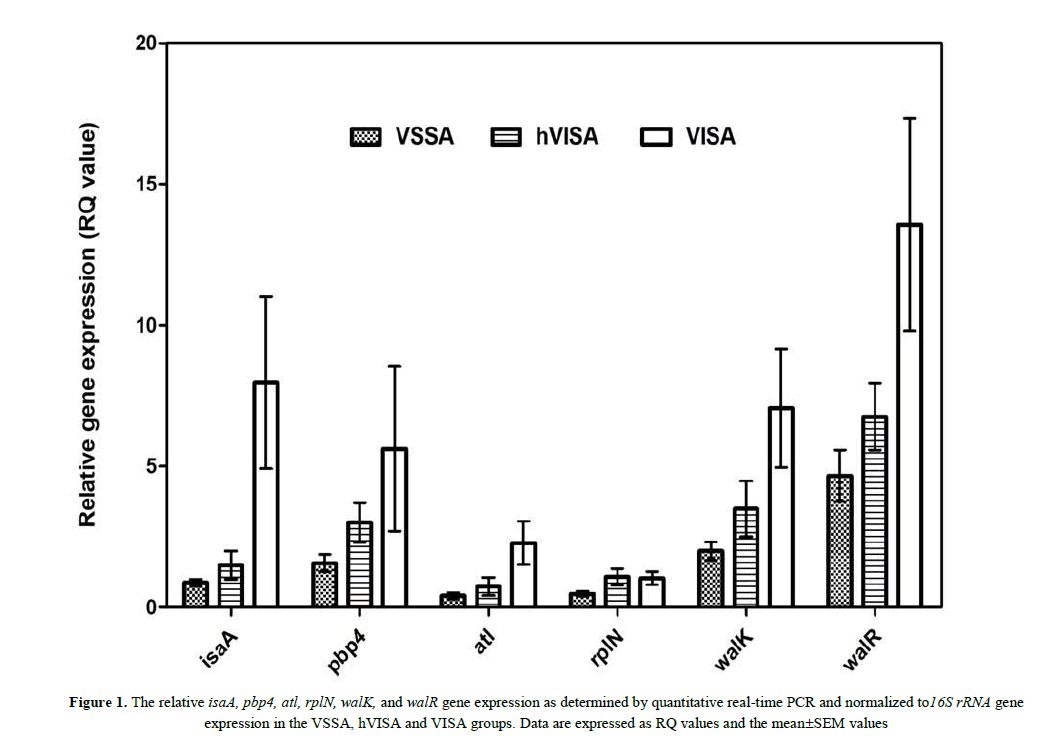
The respective expression of the six genes (i.e. isaA, atl, walK, walR, pbp4, and rplN) among the VSSA, hVISA and VISA isolates was determined by using real-time RT-PCR and displayed as RQ value. The mean±SEM of RQ values of all six genes and the fold-increase in the mean RQ values among S. aureus groups were shown in Table 3. The mean RQ values for all of the tested genes in the VISA groups were significantly higher than those of the VSSA and hVISA groups except for rplN in both the VSSA and hVISA groups and pbp4 in the hVISA group. The respective mean RQ values of isaA and atl expression in the VISA group were 9.1- and 5.6-fold of that in the VSSA and 5.3- and 3.1-fold of that in the hVISA. The respective mean RQ values of walK, walR, and pbp4 in the VISA group was 2- to 3-fold of those in the VSSA and hVISA groups. However, no significant difference was observed in the mean RQ value of rplN among the three groups and that of pbp4 in the VISA and hVISA groups. Upon comparing the mean RQ values for all six genes of hVISA with those of VSSA, no significant difference was found. Figure 1 presents the relative gene expression as the RQ value of the six genes among the three bacterial groups. In addition, it should be noted that no correlation of the expression levels of the studied genes with neither MIC nor SCCmec type among the strains in three groups (data were not shown).
| Gene | RQ value (mean ± SEM) | Fold increase (p-value)* | ||||
|---|---|---|---|---|---|---|
| VSSA (n=26) |
hVISA (n=28) |
VISA (n=15) |
VISA vs hVISA | VISA vs VSSA | hVISA vs VSSA | |
| isaA | 0.875 ± 0.123 | 1.492 ± 0.500 | 7.956 ± 3.044 | 5.3 (0.001) |
9.1 (< 0.001) |
1.7 (0.692) |
| atl | 0.407 ± 0.117 | 0.743 ± 0.322 | 2.281 ± 0.769 | 3.1 (0.009) |
5.6 (0.002) |
1.8 (0.494) |
| rplN | 0.478 ± 0.096 | 1.077 ± 0.291 | 1.030 ± 0.233 | 0.9 (0.890) |
2.2 (0.130) |
2.3 (0.052) |
| walK | 1.994 ± 0.318 | 3.491 ± 0.988 | 7.057 ± 2.097 | 2.0 (0.033) |
3.5 (0.003) |
1.8 (0.286) |
| walR | 4.665 ± 0.912 | 6.756 ± 1.183 | 13.567 ± 3.765 | 2.0 (0.013) |
2.9 (0.002) |
1.5 (0.360) |
| pbp4 | 1.566 ± 0.308 | 3.004 ± 0.703 | 5.616 ± 2.922 | 1.9 (0.165) |
3.6 (0.035) |
1.9 (0.367) |
n = Number of isolates
p< 0.05 = Significant difference
Table 3: Comparison of the mean of relative expression of six genes expressed as RQ value in the VISA, hVISA and VSSA groups
Using a cut-off value based on an RQ value above the mean + 2 SEMs of the VSSA group, we found more than half of the VISA isolates for all genes fell into this group (except for pbp4). Six, 10, and 11 of the 15 VISA isolates (40%, 66.7%, and 73.3%) had RQ values for pbp4, rplN, and walR above the cut-off values respectively. Nine VISA isolates (60%) had RQ values for isaA, atl, and walK above the cut-off values. In contrast, less than half of the hVISA isolates had RQ values for all the genes above the cut-off values. Based on the cut-off values for the six genes, 13 of 15 (86.7%) VISA and 17 of 26 (60.7%) hVISA isolates had at least one gene with an RQ value above the RQ cut-off value.
Discussion
The definite molecular pathway of hVISA/VISA occurrence from VSSA remains unclear. It has been reported that MRSA strains with low level vancomycin-resistance have undergone metabolic changes; such as, slowed growth rate, increased autolytic activity, and decreased agr expression (Lina et al., 2003). In addition, reduced the function of the agr operon leads to the reduced RNAIII expression and a reduction of certain virulence gene (s) (i.e.,spa, hla), (Sieradzki and Tomasz, 2003; Peleg et al., 2009) which facilitate bacterial evasion of the host immune system (Boisset et al., 2007). Point mutations, moreover, were found on the regulatory genes walKR and rpoB and other effector genes among the induced and natural VISA strains (Ruef, 2004; Gardete et al., 2012). The present study documented a significant difference in the mean expression of isaA and atl in VISA which were 5.3- and 3.1-fold of that in hVISA, and 9.1- and 5.6-fold of that in the VSSA. The significant increase in isaA expression in the VISA but not significant between hVISA and VSSA strains was partially consistent with a previous report that found an up-regulation of the IsaA protein in hVISA compared with the isogenic VSSA from the same patient (Chen et al., 2013). IsaA is a lytic transglycosylase enzyme which hydrolyzes peptidoglycan allowing cell growth, division, (Stapletonet al., 2007) including septation, while Atl—the most predominant peptidoglycan hydrolase in staphylococci with an amidase domain and a glucosaminidase domain—functions as an autolysin, degrading the peptidoglycan cell wall layer (Vidaillac et al., 2013).
This bifunctional hydrolase plays a key role in (a) bacterial cell wall metabolism, (b) daughter-cell separation, and (c) antibiotic mediated cell lysis (Vollmer et al., 2008). Increasing atl expression possibly reflects an increase in cell wall turnover rates. The increase of atl expression in VISA and hVISA strains in the current study is similar to a previous report in which was found an abundant increase of peptidoglycan hydrolase and transglycosylase protein in the cell envelope fraction of a VISA strain compared to the isogenic hVISA and VSSA from the same patient (Ramadurai et al., 1999). Notwithstanding, Utaida et al. (2006) reported a reduction in the whole-cell autolytic activity of the VISA strain Mu50. The discrepancy may be due to the different approach used in their study and/or to the complexity of cell wall synthesis/remodeling autolytic regulation. Such findings, however, imply that VSSA may develop into hVISA and VISA via differentiation through various metabolic pathways. In the current study, rplN coding for RplN protein—which functions as a peptidyltransferase forming peptide bonds between adjacent amino acids (Garret and Grisham, 2002)—showed no significant difference in mean rplN expression among the three groups. In our previous proteomic study, we found that VISA group had higher RplN expression than the isogenic hVISA and VSSA groups (Sirichoat et al., 2016). The discordance results of rplN expression in both studies may be due to the difference of study technique (gene expression vs. protein expression) and the phase of bacteria used in the studies (exponential vs. stationary phase). The histone-like protein (Hup) and RplN are important for optimal survival of bacterial cells in the stationary phase and under stress conditions (Balandina et al., 2002).
PBP4 of S. aureus is a carboxypeptidase needed for the secondary cross-linking of peptidoglycan (Henze and Berger-Bachi, 1995). It is a member of the enzyme group for synthesis and modification of cell wall. PBP4 overexpression causes an increase in β-lactam resistance and greater cross-linking of the peptidoglycan (Henze and Berger-Bachi, 1996). In the present study, the VISA and hVISA had a 3.6- and 1.9-fold increase in pbp4 expression over that of the VSSA. This finding agrees with a previous study that reported cell wall thickening of VISA and hVISA, which may be the result of increasing expression of pbp4 (Hiramatsu, 2001). This is in agreement with the finding that PBP4 activity was related to peptidoglycan cross-linking (Piepe et al., 2006) which might be involved in cell wall thickening. Sieradzkiet al. (1999) however, found a reduction in pbp4 activity in the VISA strain from a patient with a persistent infection.
The walRK—a two-component regulatory system—is a key regulator of cell wall metabolism in S. aureus (Atilino et al., 2010) and different point mutations in these genes were frequently found among hVISA/VISA strains (Stock et al., 2000). Although the sequences were not determined in the current study, an approximate 2- to 3.5-fold increased expression of the walRK genes was found among the VISA/hVISA strains which may have influenced the expression of other genes such as atl (Howden et al., 2011). Cameron et al. (2016) however, found YycHI mutations among VISA strains, leading to a reduction in WalRK activation. It is possible that mutations occurring in different positions within these genes may have an influence on the level of walRK expression (Stock et al., 2000). Besides the genes described above, there are a number of genes that have been reported to be associated with vancomycin resistance in S. aureus especially the VraSR operon which regulates cell wall stress stimulon was reported to up-regulate in VISA strains (Kuroda et al., 2003). However, this gene was not investigated in the present study.
Using a cut-off value based on the RQ mean+2 SEM of the VSSA group, the simultaneous increase in expression of the 6 genes was found in 1 VISA isolate; of 5 genes was found in 1 VISA; of 4 genes was found in 2 VISA and 3 hVISA; of 3 genes was found in 1 VISA and hVISA each; of 2 genes was found in 1 VISA and 3 hVISA. This finding reflects the diverse gene expression patterns among the VISA/hVISA isolates and suggests that VSSA strains are able to transform to hVISA/VISA strains through various diverse genetic pathways. In addition, mutation in the involved genes and/or their corresponding regulatory region may play a vital role in the development of hVISA/VISA strains (Howden et al., 2008). Further study is needed with larger sample sizes of VISA, hVISA, and VSSA in order to analyze for possible gene expression patterns present among these groups, which might be used for discriminating between the vancomycin-susceptible and non-susceptible S.aureus strains and/or lead to a better understanding on the intricate genetic factors involved in the development of vancomycin resistance in S. aureus.
Conclusion
In conclusion, we analyzed the expression of six genes in VISA and hVISA compared to VSSA strains and found that in particular isaA was up-regulated in VISA compared to hVISA and VSSA. Due to the complex and unknown nature of the molecular mechanism for developing VISA and hVISA, further study with more number of all strains are required to find a particular set of diagnostic marker genes to develop as a tool for fast and reliable diagnostics for effective treatment of patients with S. aureus infection.
Acknowledgments
The authors thank (a) the staff of the Clinical Microbiology Laboratory at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University, for collecting the clinical isolates, and (b) Mr. Bryan Roderick Hamman for assistance with the English-language presentation of the manuscript.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Funding
The project was supported by a research grant from Khon Kaen University (Project No. 590048) and Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences
Authors' Contributions
A.L. and V.L. designed the experiments and prepared the manuscript; A.S. performed the experiments, analyzes the data and drafts the manuscript, while A.L., A.C., R.T., and S.W. performed the experiments. All authors interpreted the data, reviewed and approved the manuscript.
About the Authors
Corresponding Author
Aroonlug Lulitanond
Faculty of Associated Medical Sciences, Centre for Research and Development of Medical Diagnostic Laboratories, Khon Kaen University, Khon Kaen, Thailand
- Email:
- arolul@kku.ac.th
References
- Atilino ML, Pereira PM, Yates J, Reed P, et al. (2010) Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci 107: 18991-18996. https://doi.org/10.1073/pnas.1004304107
- Balandina A, Kamashev D, Rouviere-Yaniv J (2002) The bacterial histone-like protein HU specifically recognizes similar structures in all nucleic acids. DNA, RNA, and their hybrids. J Biol Chem 277: 27622-27628. https://doi.org/10.1074/jbc.m201978200
- Berger-Bachi B, Barberis-Maino L, Strässle A, Kayser FH (1989) FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet 219: 263-269. https://doi.org/10.1007/bf00261186
- Boisset S, Geissmann T, huntzinger E, Fechter P, et al. (2007) Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 2: 1353-1366. https://doi.org/10.1101/gad.423507
- Cameron DR, Jiang JH, Kostoulias X, Foxwell DJ, et al. (2016) Vancomycin susceptibility in methicillin-resistant Staphylococcus aureus is mediated by YycHI activation of the WalRK essential two-component regulatory system. Sci Rep7: 30823. https://doi.org/10.1038/srep30823
- Chen H, Liu Y, Zhao C, Xiao D, et al. (2013) Comparative proteomics-based identification of genes associated with glycopeptide resistance in clinically derived heterogeneous vancomycin-intermediate Staphylococcus aureus strains. PLoS One 8: 1-10. https://doi.org/10.1371/journal.pone.0066880
- Clinical and Laboratory Standards Institute (2010) Performance standards for antimicrobial susceptibility testing. Nineteenth information supplement M100-S19, CLSI.
- Cui L, Ma X, Sato K, Okuma K, et al. (2003) Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol 41: 5-14. https://doi.org/10.1128/jcm.41.1.5-14.2003
- Cui L, Neoh HM, Shoji M, Hiramatsu K. (2009) Contribution of vraSR and graSR point mutations to vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother 53: 1231-1234. https://doi.org/10.1128/aac.01173-08
- Fridkin SK, Hageman J, McDougal LK, Mohammed J, et al. (2003) Epidemiological and microbiological characterization of infections caused by Staphylococcus aureus with reduced susceptibility to vancomycin, United States, 1997-2001. Clin Infect Dis 36: 429-439. https://doi.org/10.1086/346207
- Gardete S, Kim C, Hartmann BM, Mwangi M, et al. (2012) Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog 8: e1002505. https://doi.org/10.1371/journal.ppat.1002505
- Garret R and Grisham C (2002) Mechanisms of enzyme action. In: Berg JM, Tymoczko JL, Stryer L, editors. Biochemistry. (5th edn).WH Freeman, New York, USA.
- Henze UU and Berger-Bachi B (1995) Staphylococcus aureus penicillin-binding protein 4 and intrinsic beta-lactam resistance. Antimicrob. Agents Chemother 39: 2415-2422. https://doi.org/10.1128/aac.39.11.2415
- Henze UU and Berger-Bachi B (1996) Penicillin-binding protein 4 overproduction increases beta-lactam resistance in Staphylococcus aureus. Antimicrob. Agents Chemother 40: 2121-2125. https://doi.org/10.1128/aac.40.9.2121
- Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, et al. (1997) Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350: 1670-1673. https://doi.org/10.1016/s0140-6736 (97)07324-8
- Hiramatsu K (2001) Vancomycin-resistant Staphylococcus aureus: A new model of antibiotic resistance. Lancet Infect Dis 1: 147-155. https://doi.org/10.1016/s1473-3099 (01)00091-3
- Howden BP, Smith DJ, Mansell A, Johnson PD, et al. (2008) Different bacterial gene expression patterns and attenuated host immune responses are associated with the evolution of low-level vancomycin resistance during persistent methicillin-resistant Staphylococcus aureus bacteraemia. BMC Microbiol 8: 39. https://doi.org/10.1186/1471-2180-8-39
- Howden BP, Davies JK, Johnson PD, Stinear TP, et al. (2010) Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev 23: 99-139. https://doi.org/10.1128/cmr.00042-09
- Howden BP, McEvoy CRE, Allen DL, Chua K, et al. (2011) Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS. Pathog 7: e1002359. https://doi.org/10.1371/journal.ppat.1002359
- Kondo Y, Ito T, Ma XX, Watanabe S, et al. (2007) Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother 51: 264-274. https://doi.org/10.1128/aac.00165-06
- Kuroda M, Kuroda H, Oshima T, Takeuchi F, et al. (2003) Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49: 807-821. https://doi.org/10.1046/j.1365-2958.2003.03599.x
- Lina G, Boutite F, Tristan A, Bes M, et al. (2003) Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl. Environ. Microbiol 69: 18-23. https://doi.org/10.1128/aem.69.1.18-23.2003
- Liu C and Chambers HF (2003) Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother 47: 3040-3045. https://doi.org/10.1128/aac.47.10.3040-3045.2003
- Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods 25: 402-408. https://doi.org/10.1006/meth.2001.1262
- Matsuo M, Hishinuma T, Katayama Y, Cui L, et al. (2011) Mutation of RNA polymerase beta subunit (rpoB) promotes hVISA- to -VISA phenotypic conversion of strain Mu3. Antimicrob. Agents Chemother 55: 4188-4195. https://doi.org/10.1128/aac.00398-11
- Mwangi MM, Wu SW, Zhou Y, Sieradzki K, et al. (2007) Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci 104: 9451-9456. https://doi.org/10.1073/pnas.0609839104
- Peleg AY, Monga D, Pillai S, Mylonakis E, et al. (2009). Reduced susceptibility to vancomycin influences pathogenicity in Staphylococcus aureus infection. J Infect Dis 199: 532-536. https://doi.org/10.1086/596511
- Piepe R, Gatlin-Bunai CL, Mongodin EF, Parmar PP, et al. (2006) Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 6: 4246-4258. https://doi.org/10.1002/pmic.200500764
- Ramadurai L, Lockwood KJ, Nadakavukaren MJ, Jayaswal RK (1999) Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiol 145: 801-808. https://doi.org/10.1099/13500872-145-4-801
- Ruef C (2004) Epidemiology and clinical impact of glycopeptide resistance in Staphylococcus aureus. Infection.32: 315-327. https://doi.org/10.1007/s15010-004-4124-7
- Satola SW, Farley MM, Anderson KF, Patel JB (2011) Comparison of detection methods for heteroresistant vancomycin-intermediate Staphylococcus aureus, with population analysis profile method as the reference method. J Clin Microbiol 49: 1177-1183. https://doi.org/10.1128/jcm.01128-10
- Shittu A, Lin J, Morrison D, Kolawole D (2004) Isolation and molecular characterization of multiresistant Staphylococcus sciuri and Staphylococcus haemolyticus associated with skin and soft-tissue infections. J Med Microbiol 53: 51-55. https://doi.org/10.1099/jmm.0.05294-0
- Sieradzki K, Roberts RB, Serur D, Hargrave J, et al. (1999) Heterogeneously vancomycin-resistant Staphylococcus epidermidis strain causing recurrent peritonitis in a dialysis patient during vancomycin therapy. J Clin Microbiol 37: 39-44.
- Sieradzki K and Tomasz A (2003) Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J Bacteriol 185: 7103-7110. https://doi.org/10.1128/jb.185.24.7103-7110.2003
- Sirichoat A, Lulitanond A, Kanlaya R, Tavichakorntrakool R, et al. (2016) Phenotypic characteristics and comparative proteomics of Staphylococcus aureus strains with different vancomycin-resistance levels. Diagn. Microbiol. Infect Dis 86: 340-344. https://doi.org/10.1016/j.diagmicrobio.2016.09.011
- Stapleton MR, Horsburgh MJ, Emma J, Wright L, et al. (2007) Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol 189: 7316-7325. https://doi.org/10.1128/jb.00734-07
- Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Ann Rew Biochem 69: 183-215. https://doi.org/10.1146/annurev.biochem.69.1.183
- Utaida S, Pfeltz RF, Jayaswal RK, Wilkinson BJ (2006) Autolytic properties of glycopeptides intermediate Staphylococcus aureus Mu50. Antimicrob. Agents Chemother 50: 1541-1545. https://doi.org/10.1128/aac.50.4.1541-1545.2006
- Vidaillac C, Gardete S, Tewhey R, Sakoulas G, et al. (2013) Alternative mutational pathways to intermediate resistance to vancomycin in methicillin-resistant Staphylococcus aureus. J Infect Dis 208: 67-74. https://doi.org/10.1093/infdis/jit127
- Vollmer W, Joris B, Charlier P, Foster S (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32: 259-286. https://doi.org/10.1111/j.1574-6976.2007.00099.x
- Wootton M, Howe RA, Hillman R, Walsh TR, et al. (2001) A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 47: 399-403. https://doi.org/10.1093/jac/47.4.399
Keywords:
Download:
Full PDF- Share This