Two marine sponges-associated cultivable bacteria: Diversity and biological activities
Received: February 22, 2018
Accepted: April 24, 2018
Published: April 28, 2018
Genet.Mol.Res. 17(2): gmr16039910
DOI: 10.4238/gmr16039910
Abstract
Marine sponges harbor diverse bacterial communities. Sponge associated bacteria produce potential secondary metabolites of medical use. Little is known about sponge associated diversity from red sea therefore, we have collected two sponge samples i.e., Pione vastifica and Siphonochalinna siphonella collected from north of red sea in Obhur region, Jeddah Saudi Arabia. By using culture dependent method, we have isolated 95 different bacterial species from two marine sponge samples. These marine bacteria were screened for their antagonistic potential against fungal pathogens (Phytophthora capsici and Pythium ultimum) and human pathogenic bacteria (E. coli, Methicillin-resistant Staphylococcus aureus, E. faecalis, and P. aeroginosa). Among all 37 (39%) marine bacteria showed inhibition against oomycetes, only 27 (28.4%) exhibited antibacterial activity while 19 (20%) exhibited both antifungal and antibacterial activities. These bacterial strains were further screened for enzyme production (cellulase, protease, lipase, and amylase). Most of the strains were positive for production of lipase enzyme. These antimicrobial activities and enzyme production suggest their role in marine sponge as protecting against different marine pathogens. Taxonomic and phylogenetic analyses on the basis of 16S rRNA gene sequences showed that dominant phylum was γ-Proteobacteria. Our results highlighted that marine sponges are potential source of marine bacteria producing antimicrobial metabolites and enzymes of pharmaceutical and industrial significance.
Introduction
Due to an increase in Multi Drug Resistant (MDR) bacteria, health risk possesses to human population. Therefore, there is need for effective antimicrobial compounds, especially antibiotics effective against these resistance bacteria. Marine environment is currently a promising source of potent bioactive chemicals; especially invertebrates are known to harbor diverse microbial communities due to their filter-feeding habit and symbiosis (Zhang et al., 2015). Mutualistic relationship between marine sponges and their associated microbes is important for both of them. As sponge provide place for colonization, shelter from predator and nutrients to microbe and in turn by products of sponge are eliminated by these microbes as well as bioactive compounds excreted by these microbes help against different microbial disease (Taylor et al., 2007).
Microbial communities of marine invertebrates reported to provide biologically active compounds of biotechnological and pharmaceutical significance (Blunt et al., 2016). Marine sponges are hosts for symbiotic microorganisms and recently many bioactive compounds were isolated from these microorganisms. It has been reported that more than 15,000 natural compounds with more than 8000 new compounds had been isolated from marine invertebrate where 30% discovered from marine sponges (Koopmans et al., 2009; Brinkmann et al., 2017). Natural products derived from marine sponges are diverse in function ranging from anti-inflammatory, antiviral, antitumor, immunosuppressive to antibiotic (Imhoff et al., 2011). Most of these bioactive compounds isolated from marine sponges are in fact product of associated microbes (Khan et al., 2014).
To get knowledge of sponge and associated bacterial community’s interactions, it is important to identify, study diversity and characterize marine sponges’ associated microbial communities. Both culture dependent and culture-independent techniques reported diversity of bacterial phyla associated with marine sponges (Taylor et al., 2007; Öztürk et al., 2013) and until now 26 different phyla have been reported from marine sponges (Hentschel et al., 2002; Lee et al., 2011; Webster et al., 2001). Studies have been also performed to isolate sponge associated bacteria and screened them for the production of active metabolites (Anand et al. 2006; O’Halloran et al. 2011).
However, there is lack of investigations for bacterial association with marine sponges from region of Saudi Arabia. Therefore, the aim of present study is isolation and identification of bacteria from two selected species of marine sponges namely Pione vastificaand Siphonochalina siphonella and furthers their screening for antibacterial and antifungal activities as well as characterizes them for their enzymatic potential.
Material and Methods
Sample collection
Two sponge samples were collected by SCUBA at the depth of 20-30m from the Jeddah, Red Sea. These sponge samples were identified as Pione vastificaand Siphonochalina siphonella, by Dr. Mohsin from marine science department King Abdul-Aziz University. After collection these sponge samples were covered by seawater inside sterile ziploc plastic bag and transported immediately to the laboratory for bacterial isolation.
Isolation of bacteria from sponge samples
The sponge samples were cut into small pieces and approximately 1g of each sponge sample was finely cut into small pieces and ground with a sterile mortar and pestle. Aliquots (0.1ml) were 10-fold diluted (10-3, 10-4 and 10-5) in sterile filtered seawater (FS) and 0.1ml aliquots will be spread on to isolation media. Isolation media were half strength R2A (½ R2A), half Tryptic soy agar (½ TSA), half nutrient agar (½ NA), in sea water and marine agar (MA) using distilled water. Cyclohexamide (50µg/ml) was added to inhibit the growth of fungi. Plates were incubated at 25°C for 5-7 days. Bacterial colonies were purified by transferring onto new plates. Using 1/10 R2A medium in FS isolated strains were sub-cultured and stored at -70°C in ½ R2A broth in FS containing 15% (v/v) glycerol for further use.
DNA extraction and 16S rDNAgene analysis
For identification of isolated bacterial strains, genomic DNA was extracted, and strains were subjected to 16S rDNA gene analysis. For extraction of genomic DNA, genomic DNA extraction kit (Qiagen) was used. The 16S rDNA gene fragment of almost 1.5kb was amplified fromthe extracted bacterial DNA using primers 27F (5'-AGAGTTTGATCCTGGCTCAG-3') and 1492R (5'- GGTTACCTTGTTACGACTT -3') and amplifications were performed under PCR conditions described previously (Bibi et al., 2012). PCR products were purified using PCR purification kit (Qiagen) according to the manufacturer’s instructions and sequenced commercially (Macrogen, South Korea). Bacteria were identified after blast searches of their 16S rRNA gene sequences obtained using the EzTaxon server (https://www.ezbiocloud.net)(Kim et al., 2012). Phylogenetic status of the isolated bacteria wasconfirmed using CLUSTALX (Thompson et al., 1997) and BioEdit software (Hall, 1999) was used to edit the sequences. In MEGA6 Programme, neighbor-joining method with bootstrap values based on 1,000 replications was used for construction of the phylogenetic tree based on the 16S rRNA gene sequences (Tamura et al., 2013).
Analysis of antagonistic activity against oomycetes
Antifungal activity of isolated bacteria was evaluated by checking inhibition of the growth of fungal pathogens using a confrontation bioassay as described previously (Bibi et al., 2012). The plant pathogenic oomycetes Phytophthora capsici and Pythium ultimum were obtained in this laboratory and used in bioassay. Bacteria were screened for their antagonistic activity on modified PDA media using cross streak method (Bibi et al., 2012). All bacterial isolates were streaked on modified PDA media containing ½ PDA medium with ½ R2A in sea water. Mycelial disc of 6 mm of freshly cultured fungal pathogen was placed in the center of plate and bacteria were streaked perpendicular to edges of plate at 4 cm distance and incubated for 3–5 days at 28°C. Bacteria positive for antagonistic activity were checked twice and activity was evaluated by measuring the zone of inhibition around each bacterial streak.
Screening of bacteria for antibacterial activity
Bacteria isolated from marine sponges were screened for antibacterial activity using deferred antagonistic assay. Bacterial isolates from this study were grown at 28°C for 24 hrs on ½ R2A media and then overlaid with 0.1% soft agar mixed with test strains. All test strains were diluted to final concentration A600 =0.1. Plates were again incubated at 28°C for 48 hrs and the zone of inhibition was documented. The test strains of bacteria (Escherichia coli ATCC 8739, Enterococcus faecalis ATCC 29212, Enterococcus faecium ATCC 27270 and Pseudomonas aeruginosa ATCC 27853) were pregrown in LB broth at 37°C.
Evaluation of hydrolytic enzymatic activity
Bacterial isolates from two sponges were further evaluated for their hydrolytic enzyme production. To check protease activity, skim milk ½ R2A agar plates were used. Positive isolates were exhibiting protease production and made clear zone on plates. Starch media was used to check amylase production of bacterial strains. Hydrolysis of starch was seen on agar plates as a clear zone by positive strains (Kumar et al., 2012). To check lipolytic activity of strains, ½ R2A agar media supplemented with tributyrin was used. After incubation at 28°C for 48 hrs positive isolates showed clear zone around as tributyrin hydrolyszed. For cellulase activity, bacteria were streaked on CMC agar (carboxy methyl cellulose agar) media and incubated at 28°C for 48 hrs. Plates were then flooded with congo red (0.1%) solution and placed on orbital shaker for 45 min and then rinsed with 1MNaCl (Hendricks et al., 1995). Isolates active for cellulose production were observed as making clear zone on CMC agar plates.
Nucleotide sequence accession numbers
Nucleotide sequences of the bacteria isolated from sponges were deposited in the GenBank database under accession numbers KY436424–KY436445.
Results
Isolation of rhizo and endophytic bacteria from halophytes
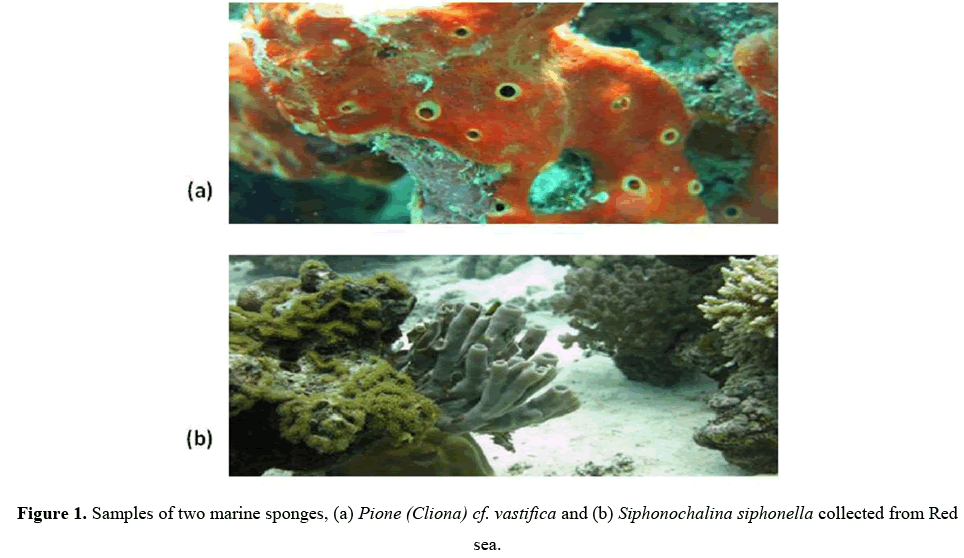
Two sponge samples, P. vastifica and S. siphonella were collected and included in this study for isolation and identification of bacteria associated with them. (Figures 1a and 1b).
Four different types of media i.e.,½ R2A, ½ TSA, ½ NA, MA were used for culturing of bacteria. Number and type of bacteria differed on different media by counting colony forming units (CFU) and looking at morphology of bacterial colony (data not shown). High numbers of bacteria were seen on ½ R2A followed by ½ TSA while low numbers of bacteria were seen on both ½ NA and MA indicating that ½ R2A and ½ TSA are favorable for isolation of bacteria from sponge samples. Low number of bacteria on these media may be due to high concentration of nutrients in MA and ½ NA didn’t favor growth of marine bacteria from marine sponges. For both sponge samples 20-40 times more CFU were isolated on ½ R2A as compare to other three media. Therefore, concentration and choice of culturing media for isolation of bacteria is very important to provide them with conditions optimal for their growth. A total of 95 bacteria were isolated from these two marine sponges using four different culturing media (Table 1).
| Sponges | Isolates | Antagonists | Antagonists (%) | Dominant phylum |
|---|---|---|---|---|
| Pione vastifica | 57 | 24 | 42.1 | gProteobacteria |
| Siphonochalinna Siphonella | 38 | 13 | 34.2 | gProteobacteria |
| 95 | 37 | 52 |
Table 1: Distribution of total number of bacteria and antagonistic one from two marine sponge samples.
Phylogenetic analysis of antagonistic bacteria based on 16S rRNA gene sequence
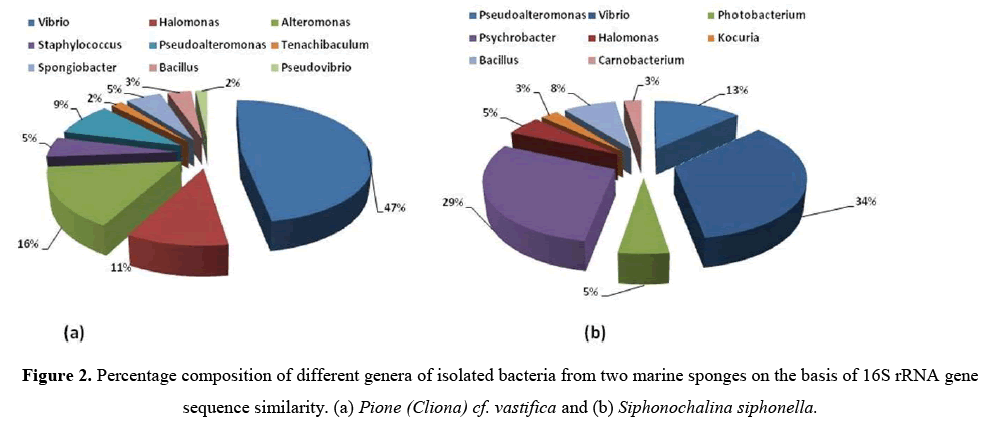
All these bacteria were identified by using 16S rRNA gene sequence analysis.From sponge P. vastifica, 9 different genera i.e.,Vibrio,Halomonas, Alteromonas, Staphylococcus,Pseudoalteromonas,Tenachibaculum, Spongiobacter, Bacillus,Pseudovibrio were identified and further belong to 3 different classes (γ-Proteobacteria, Firmicutes and Flavobacteria) where dominant class was Actinobacteria (42%) (Figure 2a).
From S. siphonella, eight different genera of bacteria were identified namely, Pseudoalteromonas,Vibrio, Photobacterium, Psychrobacter, Halomonas, Kocuria, Bacillus, Carnobacterium and belonging to three different classes (γ-Proteobacteria, Actinobacteria and Firmicutes) and dominant class was γ-Proteobacteria (34%) (Figure 2b). Sequence similarities of isolated bacteria from 94.2%–100% and 95.7%–100% from P. vastifica and S. siphonella(Table 2).
| Antifungal activityc | Antibacterial acitivityd | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lab no | Accession Number | Similarity with closest type straina | % identityb | P. capsici | P. ultimum | P.aeruginosa | S. aureus | E.coli | E.faecalis |
| P. vastifica | |||||||||
| EA250 | KY655351 | Vibrio hepatarius LMG 20362(T) | 98.8 | + | + | - | - | - | - |
| EA251 | KY655352 | Halomonas denitificans M29(T) | 98.1 | + | + | - | + | w | |
| EA252 | KY655353 | Alteromonas marina SW-47(T) | 98.7 | - | - | - | - | - | - |
| EA253 | KY655354 | Staphylococcus argenteus MSHR1132(T) | 99.8 | ++ | ++ | + | ++ | + | - |
| EA254 | KY655355 | Pseudoalteromonas shioyasakiensis SE3(T) | 99.7 | + | + | - | - | - | - |
| EA255 | KY655356 | Halomonas denitificans M29(T) | 98.2 | + | ++ | - | + | + | - |
| EA256 | KY655357 | Vibrio fortis LMG 21557 (T) | 99.7 | ++ | ++ | - | + | + | |
| EA257 | KY655358 | Vibrio harveyi ATCC 14126(T) | 99.2 | - | - | - | - | - | - |
| EA258 | KY655359 | Vibrio shilonii AK1(T) | 99.7 | + | + | - | - | - | - |
| EA259 | KY655360 | Vibrio parahaemolyticusNBRC 12711(T) | 99.5 | + | + | - | - | - | ++ |
| EA260 | KY655361 | Vibrio antiquarius Ex25(T) | 99.5 | ++ | +++ | - | w | - | ++ |
| EA261 | KY655362 | Tenachibaculum litopenaei B-1(T) | 99.5 | ++ | ++ | - | - | - | - |
| EA262 | KY655363 | Pseudoalteromonas shioyasakiensis SE3(T) | 94.3 | + | + | - | - | - | - |
| EA263 | KY655364 | Halomonas ventosae Al12(T) | 96.9 | - | - | - | - | - | - |
| EA264 | KY655365 | Pseudovibrio denitificans DN34(T) | 100 | + | + | - | - | - | + |
| EA265 | KY655366 | Halomonas ventosae Al12(T) | 98.3 | ++ | ++ | - | + | - | - |
| EA266 | KY655367 | Alteromonas marina SW-47(T) | 99.7 | - | - | + | - | + | - |
| EA267 | KY655368 | Halomonas aquamarina DMS30161(T) | 99.1 | - | - | + | - | - | - |
| EA268 | KY655369 | Alteromonas marina SW-47(T) | 94.2 | + | + | - | - | - | - |
| EA269 | KY655370 | Vibrio Caribbeanicus ATCC BAA-2122(T) | 98.6 | - | +++ | - | - | - | - |
| EA270 | KY655371 | Vibrio hepatarius LMG 20362(T) | 98.7 | - | - | - | - | - | - |
| EA271 | KY655372 | Spongiobacter nickelotolerans OOP-Ni033-1-1-2(T) | 99.5 | - | - | - | - | - | - |
| EA272 | KY655373 | Pseudoalteromonas shioyasakiensis SE3(T) | 95.7 | - | - | - | - | - | - |
| EA273 | KY655374 | Spongiobacter nickelotolerans OOP-Ni033-1-1-2(T) | 99.5 | - | - | - | - | - | - |
| EA274 | KY655375 | Alteromonas marina SW-47(T) | 93.9 | + | + | - | - | - | - |
| EA275 | KY655376 | Alteromonas macleodii ATCC 27126(T) | 99.8 | + | + | - | - | - | - |
| EA276 | KY655377 | Spongiobacter nickelotolerans OOP-Ni033-1-1-2(T) | 98.8 | ++ | ++ | - | - | - | - |
| EA277 | KY655378 | Alteromonas marina SW-47(T) | 99.1 | + | + | - | - | + | - |
| EA278 | KY655379 | Bacillus pumilus ATCC 7061(T) | 99.8 | - | - | - | w | ++ | - |
| EA279 | KY655380 | Vibrio antiquarius Ex25(T) | 99.7 | - | - | - | - | ++ | - |
| EA280 | KY655381 | Vibrio alginolyticus NBRC 15630(T) | 99.5 | - | - | - | - | - | - |
| EA281 | KY655382 | Vibrio OwensiiLMG 25443(T) | 99.8 | - | - | - | - | - | - |
| EA282 | KY655383 | Vibrio maritimus R-40493(T) | 99.7 | - | - | w | - | ++ | - |
| EA283 | KY655384 | Vibrio neocaledonicus NC470(T) | 100 | - | - | - | - | - | - |
| EA284 | KY655385 | Vibrio hepatarius LNG 20362(T) | 98.7 | + | + | - | - | ++ | - |
| EA285 | KY655386 | Vibrio Caribbeanicus ATCC BAA-2122(T) | 96.8 | - | - | - | - | - | - |
| EA286 | KY655387 | Vibrio madracius R-40493(T) | 95.6 | - | - | - | - | - | - |
| EA287 | KY655388 | Vibrio fortis LMG 21557 (T) | 100 | - | - | - | - | - | - |
| EA288 | KY655389 | Vibrio azurius NBRC 15630(T) | 99.7 | - | - | - | - | - | - |
| EA289 | KY655390 | Alteromonas gracilis 9a2(T) | 94.9 | - | - | - | - | - | - |
| EA290 | KY655391 | Vibrio madracius R-40493(T) | 99.4 | - | - | - | - | - | - |
| EA291 | KY655392 | Staphylococcus argenteus MSHR1132(T) | 100 | - | - | - | - | - | - |
| EA292 | KY655393 | Vibrio shilonii AK1(T) | 99.7 | - | - | - | - | - | - |
| EA293 | KY655394 | Alteromonas macleodii ATCC 27126(T) | 99.4 | - | - | - | - | - | - |
| EA294 | KY655395 | Staphylococcus argenteus MSHR1132(T) | 99.7 | - | - | - | - | - | - |
| EA295 | KY655396 | Vibrio hepatarius LNG 20362(T) | 98.8 | - | - | - | - | - | - |
| EA296 | KY655397 | Alteromonas macleodii ATCC 27126(T) | 99.7 | - | - | - | - | - | - |
| EA297 | KY655398 | Vibrio caribbeanicus ATCC BAA-2122(T) | 98.6 | - | - | - | - | - | - |
| EA298 | KY655399 | Halomonas meridiana DSM5425(T) | 99 | - | - | - | - | - | - |
| EA299 | KY655400 | Pseudoalteromonas shioyasakiensis SE3(T) | 99.4 | + | + | - | - | - | - |
| EA300 | KY655401 | Vibrio mediterranei CIP 103203(T) | 99.4 | - | - | - | - | - | - |
| EA301 | KY655402 | Bacillus licheniformis ATCC 14580(T) | 99.4 | ++ | +++ | - | + | - | - |
| EA302 | KY655403 | Vibrio alginolyticus NBRC 15630(T) | 99.7 | - | - | - | - | - | - |
| EA303 | KY655404 | Vibrio neocaledonicus NC470(T) | 99.7 | + | + | - | - | - | - |
| EA304 | KY655405 | Pseudoalteromonas shioyasakiensis SE3(T) | 98.4 | - | - | - | - | - | - |
| EA305 | KY655406 | Vibrio Caribbeanicus ATCC BAA-2122(T) | 98.7 | + | + | - | - | - | - |
| EA306 | KY655407 | Vibrio nereis ATCC 25917(T) | 98.5 | - | - | - | - | - | - |
| S. siphonella | |||||||||
| EA307 | KY655408 | Pseudoalteromonas shioyasakiensisSE3(T) | 100 | - | - | - | - | - | - |
| EA308 | KY655409 | Vibrio harveyi NBRC 15634(T) | 100 | - | - | - | - | - | - |
| EA309 | KY655410 | Photobacterium angustum ATCC 25915(T) | 98.8 | - | - | - | - | - | - |
| EA310 | KY655411 | Vibrio owensii LMG 25443(T) | 99.9 | - | - | - | - | - | - |
| EA311 | KY655412 | Pseudoalteromonas shioyasakiensis SE3(T) | 100 | - | - | - | - | - | - |
| EA312 | KY655413 | Psychrobacter alimentarius JG-100(T) | 99.9 | - | - | - | - | - | - |
| EA313 | KY655414 | Pseudoalteromonas shioyasakiensis SE3 (T) | 98 | - | - | - | - | - | - |
| EA314 | KY655415 | Halomonas aquamarina DSM 30161(T) | 99.9 | +++ | ++++ | - | + | - | - |
| EA315 | KY655416 | Vibrio neocaledonicus.NC470(T) | 99.9 | - | - | - | - | - | - |
| EA316 | KY655417 | Kocuria flava.HO-9041(T) | 99.8 | - | - | - | - | - | - |
| EA317 | KY655418 | Vibrio thalassae.MD16(T) | 98.8 | - | - | - | - | - | - |
| EA318 | KY655419 | Psychrobacter alimentarius JG-100 (T) | 99.7 | - | - | - | - | - | - |
| EA319 | KY655420 | Vibrio neocaledonicus NC470(T) | 99.9 | + | + | - | - | ||
| EA320 | KY655421 | Vibrio campbellii. CAIM 519 (T) | 100 | w | +++ | + | - | - | - |
| EA321 | KY655422 | Vibrio alginolyticus.NBRC 15630 (T) | 99.7 | - | - | - | - | - | |
| EA322 | KY655423 | Psychrobacter alimentarius.JG-100 (T) | 99.9 | - | - | + | + | + | |
| EA323 | KY655424 | Vibrio azureus.NBRC 104587(T) | 98.4 | + | +++ | - | - | - | - |
| EA324 | KY655425 | Pseudoalteromonas shioyasakiensisSE3(T) | 99.3 | ++++ | ++++ | - | + | - | - |
| EA325 | KY655426 | Bacillus licheniformis.ATCC14580 (T) | 98 | - | - | - | - | - | - |
| EA326 | KY655427 | Photobacterium angustum.ATCC25915(T) | 99.1 | - | - | - | - | - | - |
| EA327 | KY655428 | Pseudoalteromonas shioyasakiensis SE3(T) | 99.7 | - | - | - | - | - | - |
| EA328 | KY655429 | Bacillus safensis. FO-36b(T) | 99.2 | - | - | - | - | - | - |
| EA329 | KY655430 | Psychrobacter aquaticus CMS56(T) | 99.9 | ++++ | ++++ | - | + | - | - |
| EA330 | KY655431 | Psychrobacter alimentarius JG-100(T) | 99.9 | w | ++ | - | - | - | - |
| EA331 | KY655432 | Halomonas sulfidaeris.ATCC BAA-803(T) | 95.7 | ++ | + | - | + | + | |
| EA332 | KY655433 | Carnobacterium inhibens subsp. InhibensDSM 13024(T) | 99.7 | ++ | + | - | + | + | + |
| EA333 | KY655434 | Psychrobacter alimentarius.JG-100 (T) | 99.3 | - | - | - | - | - | - |
| EA334 | KY655435 | Bacillus subtilis subsp. Inaquosorum KCTC 13429 (T) | 100 | +++++ | ++++ | - | - | - | - |
| EA335 | KY655436 | Psychrobacter alimentarius JG-100 (T) | 98.6 | + | + | - | - | - | - |
| EA336 | KY655437 | Psychrobacter aquaticus CMS56(T) | 99.9 | - | - | - | - | - | - |
| EA337 | KY655438 | Vibrio sagamiensis LC2-047(T) | 98.5 | ++ | ++++ | - | - | ++ | - |
| EA338 | KY655439 | Psychrobacter alimentarius JG-100 (T) | 99.8 | ++ | ++ | - | - | + | - |
| EA339 | KY655440 | Vibrio azureus.NBRC 104587(T) | 98.4 | - | - | - | - | + | - |
| EA340 | KY655441 | Psychrobacter alimentarius JG-100(T) | 99.9 | - | - | - | - | - | - |
| EA341 | KY655442 | Vibrio sagamiensis LC2-047(T) | 98.5 | ++ | ++++ | - | - | - | - |
| EA342 | KY655443 | Vibrio thalassae MD16(T) | 98.1 | + | ++ | - | - | - | - |
| EA343 | KY655444 | Vibrio thalassae MD16(T) | 97.9 | - | - | + | - | + | |
| EA344 | KY655445 | Psychrobacter aquaticus CMS56 (T) | 99.5 | - | - | - | - | - | - |
Note: aIdentification based on partial 16S rRNA gene sequence analyses of all antagonistic bacteria. b% similarity with closely related type strain cAntagonistic activity of all bacteria isolated in this study. The activity was measured after 3-5 days incubation at 28°C by measuring the clear zone of mycelial growth inhibition: w; weak, -, Negative; +, 3 mm; ++, between 4 to 6 mm; +++, between 7 to 9 mm; ++++, between 10 to 12 +++++, between 13 to 15.
Table 2: Taxonomic identification, antifungal and antibacterial activity of bacteria from sponges, P. vastifica and S. Siphonella
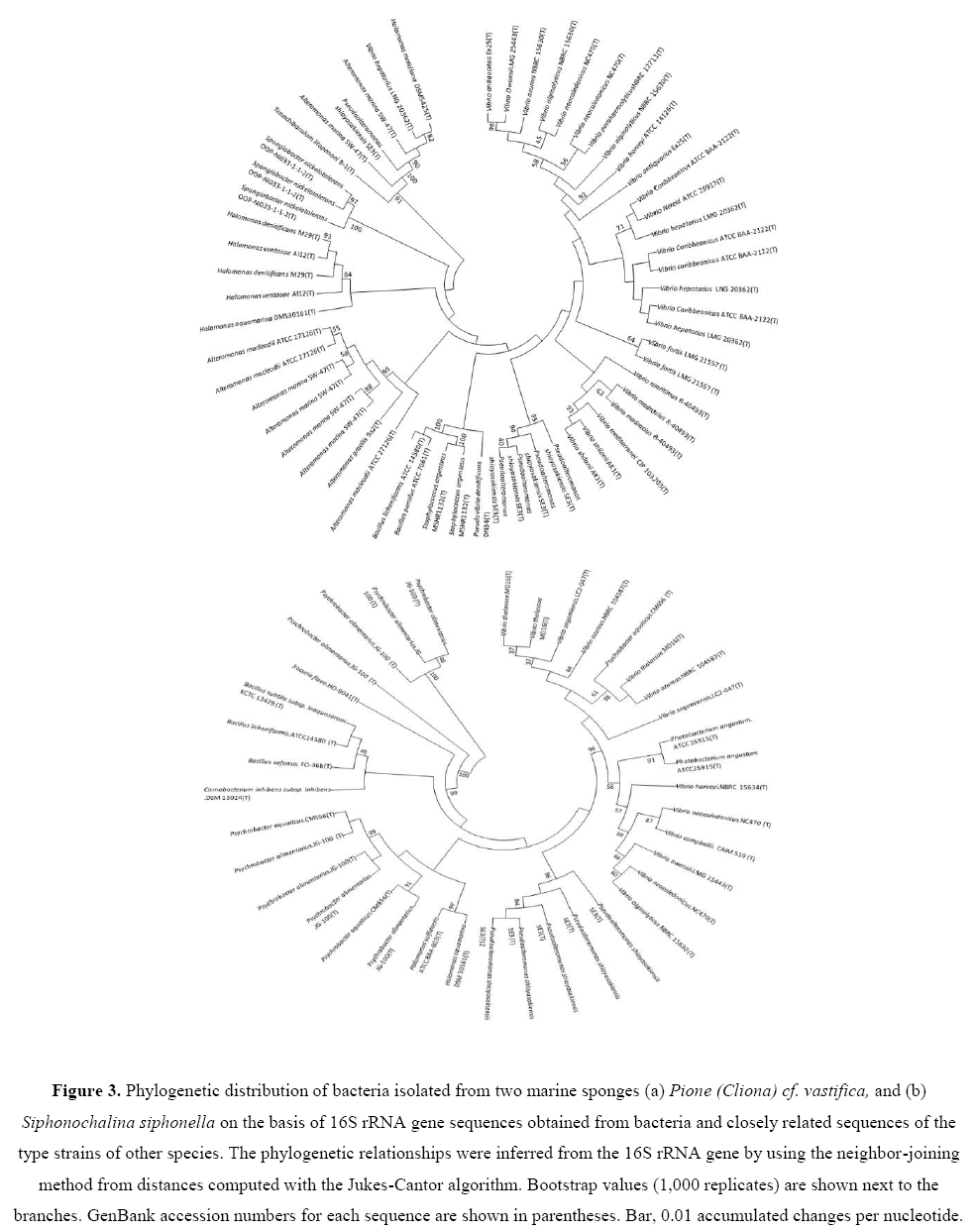
Some new and novel strains were also recovered from three marine sponges studied. Two strains Pseudoalteromonas sp. (EA262)and Alteromonas sp. (EA274) isolated from P. vastifica, were novel strain showing low 16S rRNA gene sequence similarities (93% to 94%) with respective type strains.Using 16S rRNA gene sequence data, neighbor Joining (NJ) phylogenetic trees were constructed for bacterial isolates of two marine sponges (Figures 3a and 3b). Bootstrap values were high in all three phylogenetic trees. Bacterial isolates with antagonistic activity were recovered with high bootstraps values.
Figure 3: Phylogenetic distribution of bacteria isolated from two marine sponges (a) Pione (Cliona) cf. vastifica, and (b) Siphonochalina siphonella on the basis of 16S rRNA gene sequences obtained from bacteria and closely related sequences of the type strains of other species. The phylogenetic relationships were inferred from the 16S rRNA gene by using the neighbor-joining method from distances computed with the Jukes-Cantor algorithm. Bootstrap values (1,000 replicates) are shown next to the branches. GenBank accession numbers for each sequence are shown in parentheses. Bar, 0.01 accumulated changes per nucleotide.
Antagonistic activity
Bacteria isolated from marine sponges were screened for antagonistic activity against oomycetes, Py. ultimum and P. capsici and human pathogenic bacteria, P. aeruginosa, S. aureus, E. coli and E. faecalis. From sponge P. vastifica, only 24 (42%) from 57 bacterial strains were found to be antagonistic against both oomycetes. Antagonistic bacteria from P. vastifica belong to three major classes, γ-Proteobacteria (n=51; 89%), Firmicutes (n=5; 9%), and Flavobacteria(n=1; 2%). Dominant class of bacteria from P. vastifica was γ-Proteobacteria. Sponge S. siphonella exhibited presence of 13 (34%) from total 38 bacteria isolated. Antagonistic bacteria from this sponge belong to two major classes, γ-Proteobacteria (n=33; 87%) and Firmicutes (n=5; 13%) and dominat class was also γ-Proteobacteria.Bacterial strains from marine sponges were also screened for their antibacterial activity by using agar spot test. From sponge P. vastifica, twenty bacteria (35%) showed antibacterial activity. From these strains 4 strains were active against P. aeruginosa, 12 against S. aureus, 10 against E. coli and 3 against E. faecalis. From P. vastifica, one of the strain EA253 belong to Staphylococcus sp. showed antagonistic activity against oomycetes as well as against P. aeruginosa, S. aureus, and E. coli. Proportion of bacteria exhibiting antibacterial activity was high in S. siphonella. Table 2 showed11 strains isolated from this sponge were active against different human pathogenic bacteria. Strain EA332 exhibited antifungal as well as antibacterial activity against S. aureus, E. coli and E. faecalis. In total 37 (39%)bacteria were antagonistic to oomycetes pathogens and 19 (20%) were antagonistic to human pathogenic bacteria. Dominant genus in this study from two marine spongeswas vibrio as 42 different species of this genus was detected in two marine sponges.
Enzymatic activities of antagonistic bacteria
Bacteria isolated from marine sponges were evaluated for their ability for production of cell wall lytic enzymes. The isolated 124 bacterial strains were checked for protease, amylase, lipase and cellulase activities. Hydrolytic activities among the isolates from marine sponges are summarized in Table 3.
| Enzymatic activity | ||||||
|---|---|---|---|---|---|---|
| Lab no | Accession Number | Similarity with closest type strain | Protease | Lipase | Amylase | Cellualse |
| P. vastifica | ||||||
| EA250 | KY655351 | Vibrio hepatarius LMG 20362(T) | - | - | - | - |
| EA251 | KY655352 | Halomonas denitificans M29(T) | - | - | - | - |
| EA252 | KY655353 | Alteromonas marina SW-47(T) | - | - | + | - |
| EA253 | KY655354 | Staphylococcus argenteus MSHR1132(T) | - | - | - | - |
| EA254 | KY655355 | Pseudoalteromonas shioyasakiensis SE3(T) | - | + | - | - |
| EA255 | KY655356 | Halomonas denitificans M29(T) | - | - | - | - |
| EA256 | KY655357 | Vibrio fortis LMG 21557 (T) | - | - | - | - |
| EA257 | KY655358 | Vibrio harveyi ATCC 14126(T) | - | - | - | - |
| EA258 | KY655359 | Vibrio shilonii AK1(T) | - | - | - | - |
| EA259 | KY655360 | Vibrio parahaemolyticusNBRC 12711(T) | - | + | + | - |
| EA260 | KY655361 | Vibrio antiquarius Ex25(T) | - | + | + | - |
| EA261 | KY655362 | Tenachibaculum litopenaei B-1(T) | - | - | - | - |
| EA262 | KY655363 | Pseudoalteromonas shioyasakiensis SE3(T) | - | + | - | - |
| EA263 | KY655364 | Halomonas ventosae Al12(T) | - | - | - | - |
| EA264 | KY655365 | Pseudovibrio denitificans DN34(T) | - | + | - | - |
| EA265 | KY655366 | Halomonas ventosae Al12(T) | - | + | - | - |
| EA266 | KY655367 | Alteromonas marina SW-47(T) | - | + | - | - |
| EA267 | KY655368 | Halomonas aquamarina DMS30161(T) | - | - | - | - |
| EA268 | KY655369 | Alteromonas marina SW-47(T) | - | + | + | |
| EA269 | KY655370 | Vibrio Caribbeanicus ATCC BAA-2122(T) | - | + | - | - |
| EA270 | KY655371 | Vibrio hepatarius LMG 20362(T) | - | + | - | - |
| EA271 | KY655372 | Spongiobacter nickelotolerans OOP-Ni033-1-1-2(T) | - | - | - | - |
| EA272 | KY655373 | Pseudoalteromonas shioyasakiensis SE3(T) | + | |||
| EA273 | KY655374 | Spongiobacter nickelotolerans OOP-Ni033-1-1-2(T) | - | - | - | - |
| EA274 | KY655375 | Alteromonas marina SW-47(T) | - | + | ||
| EA275 | KY655376 | Alteromonas macleodii ATCC 27126(T) | - | - | + | - |
| EA276 | KY655377 | Spongiobacter nickelotolerans OOP-Ni033-1-1-2(T) | - | - | - | - |
| EA277 | KY655378 | Alteromonas marina SW-47(T) | - | + | + | |
| EA278 | KY655379 | Bacillus pumilus ATCC 7061(T) | - | - | - | - |
| EA279 | KY655380 | Vibrio antiquarius Ex25(T) | - | + | - | |
| EA280 | KY655381 | Vibrio alginolyticus NBRC 15630(T) | - | + | + | - |
| EA281 | KY655382 | Vibrio OwensiiLMG 25443(T) | - | + | - | |
| EA282 | KY655383 | Vibrio maritimus R-40493(T) | - | + | + | - |
| EA283 | KY655384 | Vibrio neocaledonicus NC470(T) | - | + | + | - |
| EA284 | KY655385 | Vibrio hepatarius LNG 20362(T) | - | - | - | - |
| EA285 | KY655386 | Vibrio Caribbeanicus ATCC BAA-2122(T) | - | - | - | - |
| EA286 | KY655387 | Vibrio madracius R-40493(T) | - | - | + | |
| EA287 | KY655388 | Vibrio fortis LMG 21557 (T) | - | - | - | - |
| EA288 | KY655389 | Vibrio azurius NBRC 15630(T) | - | - | - | - |
| EA289 | KY655390 | Alteromonas gracilis 9a2(T) | - | - | - | - |
| EA290 | KY655391 | Vibrio madracius R-40493(T) | - | - | - | - |
| EA291 | KY655392 | Staphylococcus argenteus MSHR1132(T) | - | - | - | - |
| EA292 | KY655393 | Vibrio shilonii AK1(T) | - | - | - | - |
| EA293 | KY655394 | Alteromonas macleodii ATCC 27126(T) | - | - | - | - |
| EA294 | KY655395 | Staphylococcus argenteus MSHR1132(T) | - | - | - | - |
| EA295 | KY655396 | Vibrio hepatarius LNG 20362(T) | - | - | - | - |
| EA296 | KY655397 | Alteromonas macleodii ATCC 27126(T) | - | - | - | - |
| EA297 | KY655398 | Vibrio caribbeanicus ATCC BAA-2122(T) | - | - | - | - |
| EA298 | KY655399 | Halomonas meridiana DSM5425(T) | - | - | - | - |
| EA299 | KY655400 | Pseudoalteromonas shioyasakiensis SE3(T) | - | - | - | - |
| EA300 | KY655401 | Vibrio mediterranei CIP 103203(T) | - | - | - | - |
| EA301 | KY655402 | Bacillus lichemiformis ATCC 14580(T) | - | - | - | - |
| EA302 | KY655403 | Vibrio alginolyticus NBRC 15630(T) | - | - | - | - |
| EA303 | KY655404 | vibrio neocaledonicus NC470(T) | - | - | - | - |
| EA304 | KY655405 | Pseudoalteromonas shioyasakiensis SE3(T) | - | - | - | - |
| EA305 | KY655406 | Vibrio Caribbeanicus ATCC BAA-2122(T) | - | - | - | - |
| EA306 | KY655407 | Vibrio nereis ATCC 25917(T) | - | - | - | - |
| S. Siphonella | ||||||
| EA307 | KY655408 | Pseudoalteromonas shioyasakiensis.SE3(T) | - | - | - | - |
| EA308 | KY655409 | Vibrio harveyi.NBRC 15634(T) | + | |||
| EA309 | KY655410 | Photobacterium angustum.ATCC 25915(T) | ++ | + | ||
| EA310 | KY655411 | Vibrio owensii.LMG 25443(T) | - | - | - | - |
| EA311 | KY655412 | Pseudoalteromonas shioyasakiensis.SE3(T) | - | ++ | - | - |
| EA312 | KY655413 | Psychrobacter alimentarius.JG-100(T) | - | ++ | ++ | - |
| EA313 | KY655414 | Pseudoalteromonas shioyasakiensis.SE3 (T) | - | + | - | - |
| EA314 | KY655415 | Halomonas aquamarina DSM 30161(T) | - | + | - | - |
| EA315 | KY655416 | Vibrio neocaledonicus.NC470(T) | - | - | - | - |
| EA316 | KY655417 | Kocuria flava.HO-9041(T) | - | - | - | - |
| EA317 | KY655418 | Vibrio thalassae.MD16(T) | + | |||
| EA318 | KY655419 | Psychrobacter alimentarius.JG-100 (T) | - | - | - | - |
| EA319 | KY655420 | Vibrio neocaledonicus.NC470 (T) | - | + | - | - |
| EA320 | KY655421 | Vibrio campbellii. CAIM 519 (T) | - | + | - | - |
| EA321 | KY655422 | Vibrio alginolyticus.NBRC 15630 (T) | - | + | - | - |
| EA322 | KY655423 | Psychrobacter alimentarius.JG-100 (T) | - | - | - | - |
| EA323 | KY655424 | Vibrio azureus.NBRC 104587(T) | - | + | - | - |
| EA324 | KY655425 | Pseudoalteromonas shioyasakiensis.SE3(T) | - | + | - | - |
| EA325 | KY655426 | Bacillus licheniformis.ATCC14580 (T) | - | ++ | + | - |
| EA326 | KY655427 | Photobacterium angustum.ATCC25915(T) | - | - | - | - |
| EA327 | KY655428 | Pseudoalteromonas shioyasakiensis.SE3(T) | - | - | - | - |
| EA328 | KY655429 | Bacillus safensis. FO-36b(T) | + | ++ | - | - |
| EA329 | KY655430 | Psychrobacter aquaticus.CMS56(T) | ++ | + | - | - |
| EA330 | KY655431 | Psychrobacter alimentarius.JG-100(T) | ++ | + | - | - |
| EA331 | KY655432 | Halomonas sulfidaeris.ATCC BAA-803(T) | ++ | |||
| EA332 | KY655433 | Carnobacterium inhibens subsp. Inhibens.DSM 13024(T) | - | + | - | - |
| EA333 | KY655434 | Psychrobacter alimentarius.JG-100 (T) | - | ++ | - | - |
| EA334 | KY655435 | Bacillus subtilis subsp. Inaquosorum.KCTC 13429 (T) | +++ | ++ | ++ | - |
| EA335 | KY655436 | Psychrobacter alimentarius.JG-100 (T) | - | + | - | - |
| EA336 | KY655437 | Psychrobacter aquaticus.CMS56(T) | - | - | - | - |
| EA337 | KY655438 | Vibrio sagamiensis.LC2-047(T) | - | - | - | - |
| EA338 | KY655439 | Psychrobacter alimentarius.JG-100 (T) | - | - | - | - |
| EA339 | KY655440 | Vibrio azureus.NBRC 104587(T) | - | - | - | - |
| EA340 | KY655441 | Psychrobacter alimentarius.JG-100(T) | - | - | - | - |
| EA341 | KY655442 | Vibrio sagamiensis.LC2-047(T) | ++ | ++ | ||
| EA342 | KY655443 | Vibrio thalassae.MD16(T) | - | - | - | - |
| EA343 | KY655444 | Vibrio thalassae.MD16(T) | - | + | - | - |
| EA344 | KY655445 | Psychrobacter aquaticus.CMS56 (T) | - | - | - | - |
(Note: Production of protease, amylase, lipase, and cellulase was determined by plate assay. Enzymatic activity was estimated as zone of halo formed around bacterial colonies: -, Negative; +, 3 mm; ++, between 4 to 5 mm; +++, between 6 and 7 mm.)
Table 3: Taxonomic identification, bacterial enzyme production on different enzymatic media used for culturing and further enzymatic activities.
Isolates fromP. vastifica showed activity for both lipase (n=18, 31%) and amylase (n=10, 17%) while no bacterial isolates displayed protease and cellulase activity. Number of bacteria producing hydrolyzing enzymes S. siphonella was comparatively high from other two marine sponges. Lipase producing bacteria was high (n=22, 58%) while protease (n=7, 18%) and amylase (n=3, 8%) producing bacteria were comparatively low while all isolates were negative for production of cellulase. Mostly γ-Proteobacteria and Firmicutes strains displayed high lipase and protease production. Only one strain isolated from S. siphonella belonging to Bacillus sp. (EA334) was positive for production of protease, lipase and amylase production while few strains were positive for production of one or two different enzymes. The number of bacteria exhibiting lipase activity (n=40; 42%) from two marine sponges was high as compared to other enzymatic activities.
Discussion
Several previous studies have reported significance of microbial communities from marine sponges (Koopmans et al., 2009; Brinkmann et al., 2017). Therefore, sponges can be an untapped source of microbes that can be used as source of antibiotics and bioactive compounds. The objective of this study was to isolate bacteria, identify and screen them for their potential against different fungal and bacterial pathogens. We have used four different types of media for cultivation of bacteria and the high numbers of bacteria were recovered from ½ R2A and ½ TSA indicating that these two media contents are favorable for growth of bacteria from marine sponge. ½ R2A medium showed best recoverability as it contains nutrients in low concentration and sea water added as it contains different salts hence support growth of marine symbiotic bacteria. It was reported previously that low concentration of protein and nitrogen is important to recover diverse groups of bacteria (Joint et al., 2010; Medina et al., 2017).
In current study, the 16S rRNA sequences of bacteria associated with three marine sponges, P. vastifica, S. siphonella and S. moliss were analyzed phylogenetically. Bacterial isolates from these three marine sponges belong to 25 different genera and in turn assigned to five different classes i.e.,α-Proteobacteria,ï�? ï§-Proteobacteria, Firmicutes, Flavobacteria and Actinobacteria. Dominant class of bacteria was ï§-Proteobacteria and mostly bacteria belong to genus Vibrio where twenty-three different species were identified from total isolated bacteria (Table 2). Vibrio is a genus of marine bacteria comprising of 74 species and mostly commensal with Porifera sponges (Hoffmann et al., 2010). γ-Protobacteria in this study comprised of eight different genera i.e.,Vibrio, Pseudovibrio, Halomonas, Alteromonas, Pseudoalteromonas, Spongiobacter, Photobacterium and Psychrobacter. It is previously known that from marine environment γ-Protobacteria produced the high number of bioactive metabolites (Long & Azam, 2001). Screening of bacterial isolates resulted in 56 potential strains capable to inhibit either fungal or bacterial pathogen. Highest numbers of antagonistic bacteria were recovered from marine sponge P. vastifica.Dominant class of bacteria was γ-Protobacteria comprising of 9 different genera where vibrio was dominant genus. In this study 27 different species of genus vibrio have been identified that is really interesting. Most of strains related to this genus showed inhibition either against pathogenic fungi or bacteria. No such studies including screening of bacterial isolates for antagonistic activities have been reported before from marine sponge P. vastifica. Only one study related to boring sponge, P. vastifica reported quorum-quenching (QQ) of bacterial isolates from marine sponge as well as QQ activity of sponge extract (Saurav et al., 2016). Many potential isolates were recorded for QQ activity as well as extract of sponge also showed QQ activity. Our study is first to describe diversity of culturbale bacteria from P. vastifica and their identification and screening for bioactive characteristics. γ-Protobacteria is most common cultivable group of bacteria associated with sponge or found in surrounding water (Taylor et al., 2007; Webster and Taylor, 2012). Several previous studies have reported dominance of γ-Protobacteria using high cultivation techniques (Montalvo et al., 2014; Esteves et al., 2016).
Biologically active compounds of marine vibrios have been reported as a rich source of novel biologically active metabolites (Oclarit et al., 1994; Chen et al., 2012). More than 90 different bioactive metabolites have been isolated from this class of bacteria and many were active against pathogenic bacteria. These antibacterial compound productions are important for these rhizopheric bacteria as their ecological role in sponges hence increase their abundance (Oclarit et al., 1994; Mansson et al., 2011). In sponge P. vastifica second dominant genus was Alteromonas. Species of Alteromonas are known to produce important secondary metabolites of clinical and ecological significance. A strain of Alteromonas associated with sponge Halichondria okadai produced a bioactive tetracyclic alkaloid showing both antimicrobial and cytotoxic activity (Shigemori et al., 1992). Another dominant genus in S. siphonella was Psychrobacter comprising of 11 different species. Previous studies report antibacterial activity of Psychrobacter but no antifungal activity found yet from this genus (Kennedy et al., 2010). Strain EA329 showing close 16S rRNA similarity to Psychrobacter aquaticus exhibited strong antifungal activity in our study that is first reported. Six bacterial strains from P. vastifica and only one strain from S. siphonella showed < 97% of 16S rRNA gene sequence similarity to closely related strains hence likelt to be new or novel new taxa.
Sponge-associated bacteria actively participate in hydrolysis of accumulated macromolecules inside sponge by using their extracellular enzymes. In this natural system different types of substrate are available and hydrolytic enzymes are released by bacteria to dissolve and absorb them. Nutrients released from these substrates are then utilized by sponges (Marx et al., 2007; Lee et al., 2001). In our study high percentage of lipase activity was detected in strains from both sponges. Lipases are vital biocatalysts and can be able to hydrolyze various types of substrates and are much stable in organic solvents (Karpushova et al., 2005). Amylase producing bacteria were also detected from both sponges. Amylase helps in degradation of starch molecules and has its uses in pharmaceutical as well as in textile, cosmetic, and paper industries. Sponge-associated bacteria also produced other hydrolytic enzymes such as protease, cellulase and amylase suggests that these enzymes are important for degradation of organic substrate and regenerate nutrients in surrounding environment and sponge use them as food (Shanmughapriya et al., 2009). Most of the isolates exhibiting enzymatic activities were related to γ-Protobacteria and belonging to vibrio and Psychrobacter. In our study vibrio is dominant genus among culturable bacteria from both sponges showing antimicrobial and different enzymatic activities.
Conclusion
Our data reveals that what kind of important role sponge-associated heterotrophic bacteria are playing in seawater by producing antimicrobial compounds and degradation of substrate excreting hydrolytic enzymes. This work improves our knowledge about functional role of these bacteria in marine sponges from Red sea. Future studies related to their metabolite identification and enzyme studies will elucidate importance of novel and potential strains associated with sponges.
Acknowledgment
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH)–King Abdulaziz City for Science and Technology-the Kingdom of Saudi Arabia-award number (12-BIO3106-03). The authors also, acknowledge with thanks Science and Technology Unit, King Abdulaziz University for technical support.
About the Authors
Corresponding Author
Fehmida Bibi
Special Infectious Agents Unit, King Fahd Medical Research Center, King, Abdulaziz University, Jeddah, Saudi Arabia
- Email:
- fehmeedaimran@yahoo.com
References
- Anand TP, Bhat AW, Shouche YS, Roy U, et al., (2006). Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of South East India. Microbiological Research.Microbiol Res. 161: (3): 252-62.https://doi.org/10.1016/j.micres.2005.09.002
- Bibi F, Yasir M, Song GC, Lee SY, et al. (2012). Diversity and characterization of endophytic bacteria associated with tidal flat plants and their antagonistic effects on oomycetous plant pathogens. Plant Pathol. J. 28: 20-31.https://doi. org/10.5423/PPJ.OA.06.2011.0123
- Blunt JW, Copp BR, Keyzers RA, Munro MH, et al., (2016). Marine natural products. Nat. Prod. Rep. 33: 382–431.
- Brinkmann CM, Marker A and Kurtböke D�?° (2017). An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity 21;9(4):40. https://doi.org/10.3390/d9040040
- Chen YH, Kuo J, Sung PJ, Chang YC, et al., (2012). Isolation of marine bacteria with antimicrobial activities from cultured and field-collected soft corals. World J Microbiol Biotechnol 28: 3269–3279. https://doi.org/10.1007/s11274-012-1138-7
- Esteves AIS, Amer N, Nguyen M, Thomas T (2016). Sample processing impacts the viability and cultivability of the sponge microbiome. Front Microbiol 7:499. https://doi.org/10.3389/fmicb.2016.00499
- Hall T (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95-98.
- Hendricks CW, Doyle JD and Hugley B (1995). A new solid medium for enumerating cellulose-utilizing bacteria in soil. Appl. Environ. Microbiol. 61: 2016-2019.
- Hentschel U, Hopke J, Horn M, Friedrich AB, et al., (2002). Molecular evidence for a uniform microbial community in sponges from different oceans. Applied and environmental microbiology. 1;68(9):4431-40.https://doi.org/10.1128/aem.68.9.4431-4440.2002
- Hoffmann M, Fischer M, Ottesen A, McCarthy PJ, et al., (2010). Monday SR. Population dynamics of Vibrio spp. associated with marine sponge microcosms. The ISME journal. 4(12):1608.
- Imhoff JF, Labes A and Wiese J(2011). Bio-mining the microbial treasures of the ocean: new natural products. Biotechnol. Adv. 29 (5): 468-482. https://doi.org/10.1016/j.biotechadv.2011.03.001
- Joint I, Mühling M and Querellou J. (2010).Culturing marine bacteria–an essential prerequisite for biodiscovery. Microbial biotechnology. 1: 3(5): 564-75. https://doi.org/10.1111/j.1751-7915.2010.00188.x
- Karpushova A, Brummer F, Barth, S., Lange S, et al., (2005). Cloning, recombinant expression and biochemical characterization of novel esterases from Bacillus sp associated with the marine sponge Aplysina aerophoba. Applied Microbiology and Biotechnology 67(1): 59-69. https://doi.org/10.1007/s00253-004-1780-6
- Kennedy J, Baker P, Piper C, Cotter PD, et al. (2009). Isolation and analysis of bacteria with antimicrobial activities from the marine sponge Haliclona simulans collected from Irish Waters. Mar. Biotechnol. 11(3): 384-396. https://doi.org/10.1007/s10126-008-9154-1
- Kim OS, Cho YJ, Lee K, Yoon SH, et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62: 716-721. https://doi.org/10.1099/ ijs.0.038075-0
- Khan ST, Musarrat J, Alkhedhairy AA, Kazuo S. (2014). Diversity of bacteria and polyketide synthase associated with marine sponge Haliclona sp. Ann. Microbiol. 64: 199-207. https://doi.org/10.1007/s13213-013-0652-7
- Koopmans M, Martens D, Wijffels RH. (2009) Towards commercial production of sponge medicines. Mar. Drugs 7: 787-802. https://doi.org/10.3390/md7040787
- Kumar S, Karan R, Kapoor S, S P S, et al. (2012). Screening and isolation of halophilic bacteria producing industrially important enzymes. Braz. J. Microbiol. 43: 1595-1603. https://doi.org/10.1590/S1517-83822012000400044
- Lee OO, Wang Y, Yang J, Lafi FF, et al., (2011) Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. The ISME journal. 5(4): 650. https://doi.org/10.1038/ismej.2010.165
- Lee YK, Lee JH and Lee HK (2001). Microbial symbiosis in marine sponges. Journal of Microbiology 39(4): 254-264.
- Mansson M, Gram L and Larsen TO (2011). Production of bioactive secondary metabolites by marine vibrionaceae. Mar drugs 9: 1440-1468.
- Marx JC, Collins T, D'Amico S, Feller G, et al., (2007). Cold-adapted enzymes from marine antarctic microorganisms. Marine Biotechnology. 9: (3), 293-304. https://doi.org/10.1007/s10126-006-6103-8
- Medina D, Walke JB, Gajewski Z, Becker MH, et al., (2017). Culture media and individual hosts affect the recovery of culturable bacterial diversity from amphibian skin. Frontiers in microbiology. 24:1574. https://doi.org/10.3389/fmicb.2017.01574
- Montalvo NF, Davis J, Vicente J, Pittiglio R, et al., (2014). Integration of culture-based and molecular analysis of a complex sponge-associated bacterial community. PLoS ONE 9:e90517. https://doi.org/10.1371/journal.pone.0090517
- O’Halloran JA, Barbosa TM, Morrissey JP, Kennedy J, et al., (2011). Diversity and antimicrobial activity of Pseudovibrio spp. from Irish marine sponges. Journal of applied microbiology. 1: 110(6): 1495-508. https://doi.org/10.1111/j.1365-2672.2011.05008.x
- Oclarit J, Okada H, Ohta S, Kaminura K, et al. (1994) Anti-bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibriospecies bacterium. Microbios 78: 7–16.https://doi.org/10.14711/thesis-b1136712
- Öztürk B, De Jaeger L, Smidt H, Sipkema D (2013). Culture-dependent and independent approaches for identifying novel halogenases encoded by Crambe crambe (marine sponge) microbiota. Scientific reports. 27: (3): 2780. https://doi.org/10.1038/srep02780
- Saurav K, Bar-Shalom R, Haber M, Burgsdorf I, et al., (2016). In search of alternative antibiotic drugs: Quorum-quenching activity in sponges and their bacterial isolates. Frontiers in microbiology. 5;7:416. https://doi.org/10.3389/fmicb.2016.00416
- Shanmughapriya S, Kiran GS, Selvin J, Gandhimathi R, et al., (2009). Optimization, production, and partial characterization of an alkalophilic amylase produced by sponge associated marine bacterium Halobacterium salinarum MMD047. Biotechnology and Bioprocess Engineering 14:(1): 67- 75. http://dx.doi.org/10.1007/s12257-009-1003-1.
- Shigemori H, Bae MA, Yazawa K, Sasaki T, et al., (1992) Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J Org Chem 57: 4317-4320. https://doi.org/10.1021/jo00041a053
- Tamura K, Stecher G, Peterson D, Filipski A, et al. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725-2729. https://doi.org/10.1093/molbev/mst197
- Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295–347
- Webster NS, Taylor MW. (2012). Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol 14 (2):335–346https://doi.org/10.1111/j.1462-2920.2011.02460.x
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, et al. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882.https:// doi.org/10.1093/nar/25.24.4876
- Webster NS, Watts JE, Hill RT. (2001). Detection and phylogenetic analysis of novel crenarchaeote andeuryarchaeote 16S ribosomal RNA gene sequences from a Great Barrier Reef sponge. Marine Biotechnology. 1;3(6): 600-8. https://doi.org/10.1007/s10126-001-0065-7
- Zhang D, Sun W, Feng G, Zhang F, (2015). Phylogenetic diversity of sulphate reducing Desulfovibrio associated with three South China sea spongs. Lett Appl Microbiol. 60 (5): 504-12. https://doi: 10.1111/lam.12400
Keywords:
Download:
Full PDF- Share This