Transferability of heterologous microsatellite loci between species of Euterpe genus
Received: September 09, 2017
Accepted: October 30, 2017
Published: November 02, 2017
Genet.Mol.Res. 16(4): gmr16039825
DOI: 10.4238/gmr16039825
Abstract
Euterpe precatoria, popularly known as “açaí-do-amazonas”, has agronomic, technological and economic potential and has been gaining prominence with the commercialization of its fruits. The production of this açaí-do-amazonas is based on extractivism and management practices have been recommended for sustainable collection. To recommend sustainable rates of fruit collection, it is essential to have information on the genetic variability of natural populations to monitor the recruitment and population dynamics. The objective of this study was to evaluate the transferability of microsatellite loci of E. edulis species to E. precatoria to confirm the applicability of these markers in genetic studies. Eighteen microsatellite loci developed for E. edulis were analyzed using 20 individuals from two natural populations (Formoso and Novo Segredo) of E. precatoria collected at Feijó, State of Acre, Brazil. All loci (100%) were amplified, and of these, nine (50%) were polymorphic. A total of 29 alleles were found in the Novo Segredo population, and 27 alleles were found in the Formoso population, ranging from two to five alleles per locus, with a mean of three alleles per locus. The expected heterozygosity in the Formoso population varied from 0.100 to 0.668, with an average of 0.421. In the Novo Segredo population, a variation between 0.100 and 0.710, with a mean of 0.418, was found. The observed heterozygosity values ranged from 0.100 to 0.800, with averages of 0.333 and 0.267 for Formoso and Novo Segredo, respectively. This set of markers will support further studies on the molecular characterization of the natural populations of E. precatoria and assist with the recommendation of sustainable management practices and strategies for the conservation and genetic improvement of this species.
Introduction
Euterpe precatoria Mart. is a species that is widely distributed throughout Central and North America,especially in Central and Western Amazonia (Henderson, 1995). It stands out among the several genetic resources of the Amazon region for its abundance and fruit production (Steege et al., 2013). This species presents agronomic, technological and economic potential and has been highlighted by the commercialization of its pulp in the form of “açaí juice”, which presents several nutritional qualities (Yuyama et al., 2011; Yamaguchi et al., 2015). However, fruit exploitation is done in an extractive way, which can result in a decline in the natural populations and consequently in the reduction of genetic variability. In this way, the genetic characterization of exploited populations may help in the development and recommendation of sustainable management practices and adequate conservation strategies. In addition, it may direct germplasm collection strategies for breeding programs.
Because of their genetic characteristics, microsatellites (simple sequence repeats-SSRs) are ideal markers to study the genetic variability in natural plant populations because they are highly polymorphic, multi-allelic and codominant (Kalia et al., 2011; Fortes et al., 2016). The evaluation of genetic variability in natural populations of açaí-do-amazonas by SSR markers may help develop and adequate direction for exploiting its fruits by using various management techniques. However, the use of this marker requires the previous development of loci, which do not exist for E. precatoria, and this process is laborious and expensive (Gupta et al., 2016). The conservation of microsatellite sites among related species makes it possible to transfer them to other species (Guidugli et al., 2010; Santos et al., 2011, Carmo et al., 2015, Azêvedo et al., 2016; Fagundes et al., 2016). To date, 18 SSR loci have been developed for Euterpe edulis, seven of which have been shown to be amplifiable and polymorphic for E. oleracea (Gaiotto et al., 2001; Oliveira et al., 2010).
The success of the transferability of SSR loci between species has been observed in other palm trees. Microsatellites developed for Astrocaryum aculeatum presented a transferability rate between 50% and 93% for four species of the genera (A. acaule; A. jauari; A. murumuru and A. vulgare) and for two species of different genera (Bactris gasipaes and E. precatoria) (Ramos et al., 2012). Fortes et al. (2016) obtained 75% transferability when SSR loci were developed and applied for the species of A. aculeatum and peach palm (B. gasipaes) for the genome of A. vulgaris. In a study using SSR coconut loci (Cocus nucifera) to Butia odorata,40% transferability was observed (Mistura et al., 2012). By considering the cost of developing specific markers and the advantage of using SSR markers among related species, the objective of the present study was to evaluate the transferability of microsatellite loci of E. edulis species to E. precatoria to confirm the applicability of these markers for genetic studies with E. precatoria.
Material and Methods
Plant material
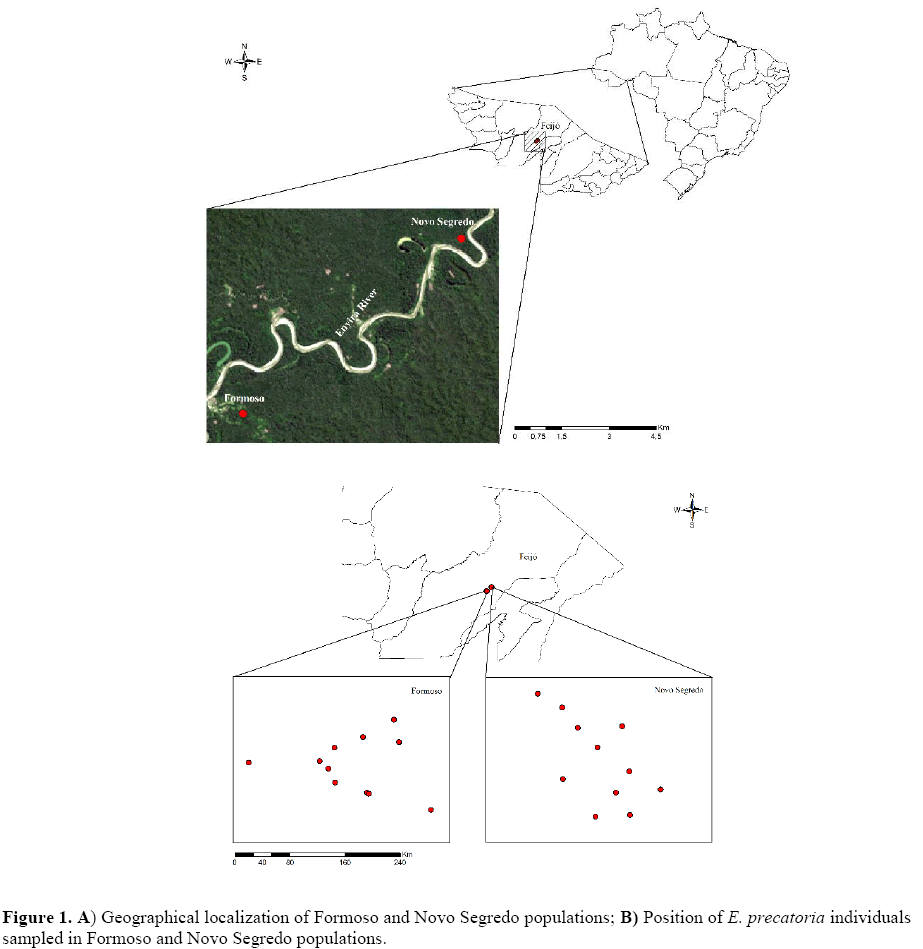
Mature leaflets were collected from 20 E. precatoria individuals from two natural populations: Novo Segredo (latitude 8S 20’31”, longitude 45 W 37’41”, 231 m above sea level) and Formoso (latitude 8S 25’40”, longitude
46 W 14’18”, 211 m above sea level). Ten individuals per population were analyzed. The populations are located in the Kaxinawá Indian Reservation of Nova Olinda, Feijó, State of Acre, Brazil (Figure 1).
DNA extraction and microsatellite loci
Total genomic DNA was extracted according to the protocol of Doyle and Doyle (1990) with modifications. They used 100 mg of leaf tissue per sample. The extracted DNA was quantified on agarose gels (1%) by comparing phage λ DNA standards that were visualized by UV light and photographed. In the amplification reactions, 18 microsatellite loci developed for E. edulis were tested (Gaiotto et al., 2001). Reactions were made containing the following: 5 ng of genomic DNA; 10 mM Tris-HCl, 50 mM KCl; 0.25 mM of each dNTP; 0.25 mg/ml BSA (Bovine Serum Albumin); 2.0 mM MgCl2; 0.2 μM of each primer; and 1 U of Taq DNA polymerase and sterile ultrapure water.
Amplifications were performed in a thermocycler (Analitik Jena), and to verify the specific annealing temperature of each locus, amplification with 12 temperature gradients (45.0°C, 45.8°C, 46.8°C, 48.2°C, 50.2°C, 52.1, 53.6, 55.4°C, 57.3°C, 58.5°C, 59.4°C and 60°C) was carried out with a sample. Loci that did not present a satisfactory standard of amplification in this range were submitted to tests at temperatures of 61°C and 62°C.
After identifying the specific annealing temperature, the samples were submitted to the following amplification steps: 94°C for 1 minute, followed by 30 cycles at 94°C for 1-minute, annealing temperature set for each primer for 1 minute and 72°C for 1 minute, followed by a final extension step at 72°C for 5 minutes. To evaluate the PCR product, the samples were submitted to agarose gel electrophoresis (3%). After amplification, the DNA fragments were separated on denaturing polyacrylamide gels (5%). Gel staining was performed using silver nitrate (Creste et al., 2001). Interpretation of amplified fragments was performed by comparison with a standard molecular weight marker (10 bp ladder - Life Technologies).
Statistical analysis
The transferability assessment was done as described by Fortes et al. (2016) based on three criteria: loci amplification, their sharpness and the lack of nonspecific bands. The polymorphic loci were characterized according to the number of alleles per locus (A), expected (HE) and observed (HO) heterozygosity, polymorphism information content (PIC) for each locus and the mean of all loci, using the TFPGA software version 1.3 (Miller, 1997). The presence of null alleles was also verified using the MICRO-CHECKER 2.2.3 software (Oosterhout et al., 2004). Gene homology were analysed by BLAST searches in the GenBank database.
Results and Discussion
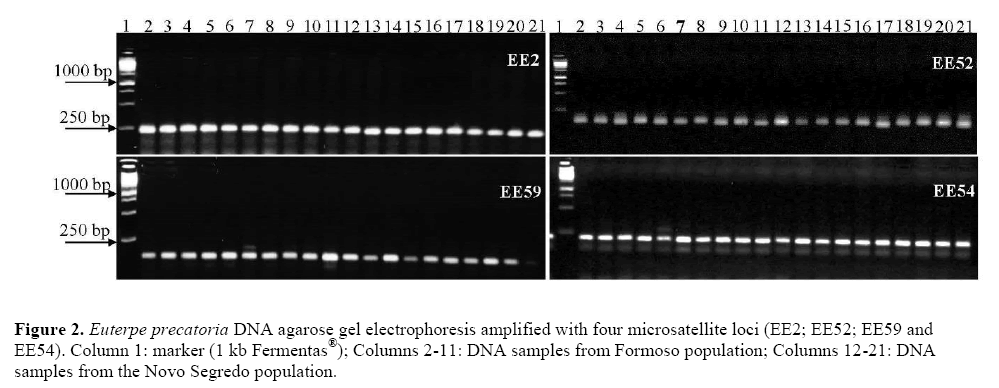
Of the 18 microsatellite loci developed for evaluated E. edulis species, 100% were amplified in the 20 analyzed genotypes of E. precatoria of Formoso and Novo Segredo populations (Table 1 and Figure 2).
| Formoso (N=10) | Novo Segredo (N=10) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | TA (°C) | Fragme size (bp) | A | HE | HO | PIC | A | HE | HO | PIC | GenBank |
| *EE2 | 55.4 | 87-90 | 2 | 0.100 | 0.100 | 0.095 | 2 | 0.100 | 0.100 | 0.095 | AF328879 |
| *EE8 | 50.2 | 120-143 | 4 | 0.647 | 0.400 | 0.615 | 4 | 0.363 | 0.200 | 0.345 | AF328872 |
| *EE32 | 55.4 | 195-215 | 5 | 0.568 | 0.500 | 0.540 | 5 | 0.710 | 0.600 | 0.675 | AF328883 |
| *EE41 | 45.8 | 124-127 | 2 | 0.189 | 0.200 | 0.180 | 4 | 0.489 | 0.200 | 0.465 | AF328884 |
| *EE45 | 57.3 | 108-110 | 4 | 0.668 | 0.400 | 0.635 | 3 | 0.694 | 0.400 | 0.660 | AF328887 |
| *EE47 | 45.8 | 270-278 | 2 | 0.505 | 0.800 | 0.480 | 4 | 0.552 | 0.300 | 0.525 | AF328874 |
| *EE52 | 53.6 | 205-210 | 3 | 0.568 | 0.200 | 0.540 | 2 | 0.336 | 0.400 | 0.320 | AF328888 |
| *EE54 | 50.2 | 114-126 | 3 | 0.278 | 0.100 | 0.265 | 3 | 0.415 | 0.100 | 0.395 | AF328876 |
| *EE59 | 50.2 | 108-122 | 2 | 0.268 | 0.300 | 0.255 | 2 | 0.100 | 0.100 | 0.095 | AF328885 |
| EE3 | 50.2 | 190 | 1 | - | - | - | 1 | - | - | - | AF328881 |
| EE5 | 46.8 | 120 | 1 | - | - | - | 1 | - | - | - | AF328882 |
| EE9 | 62.0 | 120 | 1 | - | - | - | 1 | - | - | - | AF328880 |
| EE15 | 55.4 | 140 | 1 | - | - | - | 1 | - | - | - | AF328873 |
| EE23 | 57,3 | 110 | 1 | - | - | - | 1 | - | - | - | AF328877 |
| EE25 | 55,4 | 145 | 1 | - | - | - | 1 | - | - | - | AF328878 |
| EE43 | 46,8 | 110 | 1 | - | - | - | 1 | - | - | - | AF328886 |
| EE48 | 45,8 | 240 | 1 | - | - | - | 1 | - | - | - | AF328875 |
| EE63 | 46,8 | 90 | 1 | - | - | - | 1 | - | - | - | AF328889 |
| Total | 27 | - | - | - | 29 | - | - | - | |||
| Mean | 3.0 | 0.421 | 0.333 | 0.401 | 3.2 | 0.418 | 0.267 | 0.397 | |||
*Polymorphic locus; TA (°C): Annealing temperature; bp: Base pairs; A: Total number of alleles per locus; HE: Expected heterozygosity; Ho: Observed heterozygosity; PIC: Informative content of the polymorphis
Table 1: Characteristics of 18 heterologous microsatellite markers for Euterpe precatoria
This transferability result was superior to that obtained by Gaiotto et al. (2001) when they used the same loci in E. oleracea, and amplified only seven of the 18 tested loci. Of this set of loci, ten (EE3; EE8; EE9; EE23; E25;EE32; EE41; EE45; EE47 and EE48) showed amplification with the presence of secondary products. Although the loci presented nonspecific products, all of them were analyzed to verify the presence of polymorphisms. Variations of the annealing temperature occurred among the evaluated loci (Table 1). Loci EE41 and EE48 displayed the lowest temperature values (45.8°C), and locus EE9 presented the highest annealing temperature (62.0°C). All temperatures observed in the present study differed from the original temperatures used by Gaiotto et al. (2001), which ranged from 48°C to 64°C.
A similar result was obtained by Oliveira et al. (2010), who used SSR loci of E. edulis to characterize the genetic variability of 116 accessions of E. oleracea from the collection of Embrapa Amazonia Oriental germplasm. Of the seven loci used, five were needed to optimize the annealing temperature. According to the authors, the evolutionary divergence between the species E. edulis and E. oleracea was considered low.
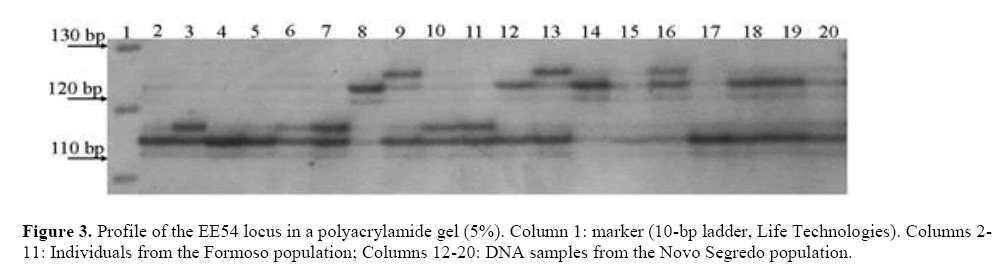
Nine loci were polymorphic (50%), ranging in size from 87 to 278 base pairs (Table 1 and Figure 3). This result was higher than that obtained by Gaiotto et al. (2001) and Oliveira et al. (2010), who tested the transferability of E. edulis markers to E. oleracea.
The total number of alleles observed in both populations was similar, with a total of 29 alleles in the Novo Segredo population and 27 alleles in the Formoso population (Table 1). The number of alleles ranged from two to five per locus, with an average of three alleles per locus. Loci EE8, EE45 and EE32 were the most polymorphic (with 4 to 5 alleles per locus) in the Formoso population, and loci EE8, EE41, and EE47 (with 4 to 5 alleles per locus) were more polymorphic in the Novo Segredo population. The loci with the lowest polymorphism in both populations were EE2 and EE59 (2 alleles per locus).
All loci exhibited less allelic variation than that observed in the species (mean = 10.6) for which they were developed (Gaiotto et al., 2001). Similar results were observed in studies with other species when transferred markers were used (Guidugli et al., 2010; Oliveira et al., 2010). The expected heterozygosity in the Formoso population ranged from 0.100 to 0.668, with a mean of 0.421. In relation to the Novo Segredo population, the variation was from 0.100 to 0.710, with a mean of 0.418. Values of observed heterozygosity ranged from 0.100 to 0.800, with means of 0.333 and 0.267 for Formoso and Novo Segredo, respectively (Table 1). The values of HOwere higher than HEat three loci in the Formoso population (EE41, EE47 and EE59) and one locus in NovoSegredo population (EE52), indicating an excess of homozygotes for these loci.
Intermediate values of HE were also observed in the population of Tucumã do Amazonas, in the municipality of Manaus, using SSR markers (HE = 0.467). The authors noted that the low value found may be related to the collection site, as the collected materials came from a forest fragment (Ramos et al., 2012). Regarding this study, the low number of heterozygotes detected in the analyzed populations may be related to the sample size since only ten individuals were analyzed in each population. Another factor that may have influenced the reduction of heterozygotes is the low number of alleles due to the locus transferability. Gaiotto et al. (2001), when using the same loci that were found to be polymorphic in the present study, verified a mean value of observed heterozygosity of 0.645. However, it must be noted that among the polymorphic loci, no locus presented null alleles.
Variations in the level of polymorphism were found among the polymorphic loci. In the Formoso population, the PIC values ranged from 0.095 to 0.635, with a mean of 0.401. In the Novo Segredo population, the values ranged from 0.095 to 0.675, with a mean of 0.377 (Table 1). The PIC values higher than 0.5 were observed in four loci in the Formoso population and in three loci in the Novo Segredo population. According to the classification defined by Botstein et al. (1980), markers with PIC values greater than 0.5 were considered highly informative.
Loci EE45 and EE59 were the two polymorphic markers that presented gene homology. There is a high probability that these loci are closely related to gene expression and may be classified as an informative functional marker. Locus EE45 presented the highest PIC value in the Formoso population (0.635) and the second highest PIC value in the Novo Segredo population (0.660). Locus EE45 showed 87% identity with a mRNA sequence of Phoenix dactylifera serine/threonine-protein kinase HT1-like (GenBank accession no. XM 008797462.2).
The EE59 locus presented homology to several sequences of MADS box transcription factor (AG1) gene in another species also of Arecaceae family. This gene family plays a deeply conserved role to the formation of sex organs in the reproductive tissue (Bowman et al., 1989).
Among the nine monomorphic loci, only one locus was homologous to known genes. The absence of polymorphism probably may be consequence of them be part of fragments of conserved genes, as the housekeeping genes. Then, they are extremely conserved in plant species. Locus EE15 presented homology to glutamate-rich WD repeat-containing protein 1-like in Elaeis guineensis and Phoenix dactylifera RNA sequences. This gene encodes a critical role in the ribosome biogenesis, another pathway highly molecular conserved. Genomic studies in Euterpe genera still don’t have released database and only extremely few sequences are available in GenBank. Our current understanding is based in 396 DNA and RNA sequences (accessed October 2017).
Conclusion
The sites conservation that flank microsatellites among related species allows the transfer of loci, and the higher the degree of relation is between species, the more efficient the transferability rates will be (Kalia et al., 2011). Hence, the transferability rates found in this study may indicate a greater phylogenetic proximity between E. edulis and E. precatoria species than found in E. oleracea (Oliveira et al., 2016). The reported microsatellitemarkers have become essential tools for genetic studies of the E. precatoria population because they play an important role in improving knowledge of the genetic variability of the species. This set of markers will support further studies of the molecular characterization of natural E. precatoria populations and assist with the definition of strategies for conservation, management and genetic improvement for this species.
Acknowledgments
The authors thank Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for granting doctorate scholarships. To colleagues Eduardo Soares and Janice Ferreira for collaboration in the figures layout.
About the Authors
References
- Azêvedo HSFS, Sousa ACB, Martins K, Oliveira JC, et al. (2016). Genetic diversity of the forage peanut in the Jequitinhonha, São Francisco, and Paranã River valleys of Brazil. Genet. Mol. Res. 15: 1-11. http://dx.doi.org/10.4238/gmr.15038601
- Botstein D, White RL, Skolnick M, Davis RW. (1980). Construction of a genetic map in man using restriction fragment length polymorphism. Am. J. Hum. Genet. 32: 314-331.
- Bowman JL, Smyth DR, Meyerowitz EM. (1989). Genes directing flower development in Arabidopsis. Plant Cell. 1: 37-52. http://dx.doi.org/10.1105/tpc.1.1.37
- Carmo FMDS, Polo EM, Silva MAD, Yazbeck GDM. (2015). Optimization of heterologous microsatellites in Piracanjuba. Pesq. Agropec. Bras. 50: 1236-1239. http://dx.10.1590/S0100-204X2015001200015
- Creste S, Tumann A, Figueira A. (2001). Detection of single sequence repeat polymorphism in denaturating polyacrylamide sequencing gels by silver staining. Plant Mol. Biol.19: 299-306. http://dx.10.1007/BF02772828
- Doyle JJ and Doyle JL. Isolation of plant DNA from fresh tissue (1990). Focus. 12: 13-15.
- Fagundes BS, Silva LF, Giacomin RM, Secco D, et al. (2016). Transferability of microsatellite markers among Myrtaceae species and their use to obtain population genetics data to help the conservation of the Brazilian Atlantic Forest. Trop. Conserv. Sci. 9: 408-422.
- Fortes ACR, Oliveira MDSP, Oliveira NP, Sanches EDNM, et al. (2016). Transferibilidade de locos microssatélites desenvolvidos em outras espécies de palmeiras para Astrocaryum vulgare Mart. Rev. Ciênc. Agrár. 59: 80-86. http://dx.doi.org/10.4322/rca.1844
- Gaiotto FA, Brondani RPV, Grattapaglia D. (2001). Microsatellite markers for heart of palm Euterpe edulis and E. oleracea Mart. (Palmae).
Mol. Ecol. Resour. 1: 86-88. - Guidugli MC, Accoroni KAG, Mestriner MA, Contel EPB, et al. (2010). Genetic characterization of 12 heterologous microsatellite markers for the giant tropical tree Cariniana legalis (Lecythidaceae). Genet. Mol. Biol. 33: 131-134.
- Gupta RK, Gupta M, Choudhury P, Mandal AB. (2016). Transferability of Rice Microsatellite Primers (SSR) Across Six Major Crops. Advances in Life Sciences. 5: 377-383.
- Henderson A (1995). The palms of the Amazon. Oxford: University Press.
- Kalia RK, Rai MK, Kalia S, Singh R, et al. (2011). Microsatellite markers: an overview of the recent progress in plants. Euphytica. 177: 309-334. http://dx.10.1007/s10681-010-0286-9
- Miller MP. (1997). Tools for population genetic analyses (TFPGA): A Windows program for the analysis of allozyme and molecular population genetic data, version 1.3. Northern Arizona University.
- Mistura CC, Barbieri RL, Castro CM, Priori D, et al. (2012). Transferibilidade de marcadores microssatélites de coco (Cocos nucifera) para butiá (Butia odorata). Magistra. 24: 360-369.
- Oliveira LC, Oliveira MDSP, Davide LC, Torres GA. (2016). Karyotype and genome size in Euterpe Mart. (Arecaceae) species. Comp. Cytogenet. 10: 17-25. http:// dx.10.3897/CompCytogen.v10i1.5522
- Oliveira MSP, Santos JB, Amorim EP, Ferreira DF, et al. (2010). Variabilidade genética entre acessos de açaizeiro utilizando marcadores microssatélites. Ciênc. Agrotec. 34: 1253-1260.
- Oosterhout CV, Hutchinson WF, Wills DPM, Shipley P. (2004). Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 4: 535-538.
- Ramos SLF, Lopes MTG, Lopes R, Cunha RNV, et al. (2012). Determination of the mating system of Tucumã palm using microsatellite markers. Crop Breed. Appl. Biotechnol. 11: 181-185. http://dx.10.3732/ajb.1100607
- Ramos SLF, Macêdo JLV, Lopes MTG, Batista JS, et al. (2012). Microsatellite loci for tucumã of amazonas (Astrocaryum aculeatum) and amplification in other Arecaceae. Am. J. Bot. 99: 508-510. http://dx.doi.org/10.3732/ajb.1100607
- Steege T, Pitman H, Sabatier NC, Baraloto DC, et al. (2013). Hyperdominance in the Amazonian tree flora. Science. 342: 1243092. http://dx.doi.org/10.1126/science.1243092
- Santos KL, Ducroquet JPHJ, Nodari RO. (2011). Caracterização genética de populações naturais de goiabeira serrana (Acca sellowiana) com marcadores microssatélites heterólogos. Biotemas. 24: 75-83. http://dx.10.5007/2175-7925.2011v24n4p75
- Yamaguchi LKK, Pereira LFR, Lamarão CV, Lima ES, et al. (2015). Amazon acai: Chemistry and biological activities: A review. Food Chem. 179: 137-151.http://dx.doi.org/10.1016/j.foodchem.2015.01.055
- Yuyama LKO, Aguiar JPL, Silva Filho DF, Yuyama K. (2011). Caracterização físico-química do suco de açaí de Euterpe precatoria Mart. oriundo de diferentes ecossistemas amazônicos. Acta Amaz. 41: 545-552. http://dx.doi.org/10.1590/S0044 59672011000400011
Keywords:
Download:
Full PDF- Share This