The clinical significance of miR-7-5p downregulation in gastric dysplasia and gastric cancer tissue
Received: October 26, 2018
Accepted: November 13, 2018
Published: January 05, 2019
Genet.Mol.Res. 18(4):
Introduction
Human gastric cancer (GC) is still the third cause of cancer-related mortality in developed countries (Tan YK and Fielding JW, 2006). As estimated GC is fifth common cancer worldwide and responsible for 984,000 new cases and 841,000 deaths were accounted in 2013 (Fitzmaurice C et al., 2015). Although managements and conventional strategies are applied for the treatment of gastric cancer, satisfactory methods have not yet found. Therefore, more investigation to seeing some new predictive and diagnostic markers are warranted, which could be helpful in finding new therapeutic strategies to improving prognosis (Yonemura Y et al., 1991; Ryu KW, Mok YJ, Kim SJ, Kim CS, 2000). Hence, studies about microRNAs might shed light on this problem.
MicroRNAs form an abundant class of short regulatory (∼21-22 nt) non-coding RNAs widely expressed in living organisms with the unique ability to negatively regulate gene expression via either translational repression or mRNA degradation (Ambros V, 2004). It has been reported that altered expression of microRNAs is associated with human diseases, particularly cancers that altered expression patterns of miRNAs could act in the oncogenic or tumor suppressor pathways (Jay C et al., 2007; Lotterman CD, Kent OA, Mendell JT, 2008). Furthermore, altered miRNA expression has been reported in various cancers including lung, pancreatic, breast and also gastric cancer (Gao W et al., 2010; Guo J et al., 2009).
miR-7 is an evolutionarily conserved miRNA. In humans, three miR-7 genes (MIR7-1,2,3) are located on chromosomes 9,15 and 19, respectively. All these three genes were giving rise to the same mature miR-7 sequence (Horsham JL et al., 2015). Several studies reported the tumor su ppressive role/down-regulation of mir-7 in pancreatic carcinoma, non-small-cell lung cancer and cervical cancer (Bi Y, Shen W, Min M, Liu Y, 2017; Xiong S et al., 2014; Hao Z et al., 2015 ).
In the present study, we investigated the expression level of miR-7 in gastric dysplasia, GC and non-tumor tissue. Such investigations could help us to develop strategies for future prevention, diagnosis and treatment of gastric cancer.
Material and Methods
Tissue samples
A total of 116 formalin-fixed paraffin-embedded (FFPE) tissue samples include 40 GC, 31 non-tumor tissue and 45 gastric dysplasia samples, were obtained from the tissue bank of Gastroenterology and Liver Disease Research Center (Tehran, Iran) between 2015 to 2017. The study procedure was approved by the organization ethical committee (IR.SBMU.MSP.REC.1396.415). Further, the experiment was conducted in complete compliance with the Declaration of Helsinki. Prepared hematoxylin and eosin stained slide of every sample were reviewed by a member of the Institute of Pathology at the of Shahid Beheshti medical university. The area of interest was marked and the slide is used to guide for scraping the unstained sections for tumor and dysplasia tissue. Non-tumor control samples were selected from normal gastric tissue resected during gastric sleeve surgery and pathology reports approved that the control samples to be free of tumor. All samples had a precise histological diagnosis consistent with gastric dysplasia and gastric cancer based on the pathology report. All data, including age, gender, lymph node metastasis (LNM), and histological grade were obtained from clinical and pathologic records. Patients that received neoadjuvant chemotherapy or radiotherapy were excluded from the study. We scraped that specified area for tumor or dysplasia enrichment. After that, total RNA extraction was performed by Qiagen miRNeasy FFPE kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol and extracted RNA subjected to DNase I treatment to remove any genomic DNA contamination. The purity and concentration of extracted RNAs were assessed using the Nanodrop ND-2000/2000c Spectrophotometer. V1.0 (Thermo Scientific).
RNA isolation and reverse transcription (cDNA synthesis)
We used poly (A)-tailed universal reverse transcription method as described elsewhere (Design R, 2005) for cDNA synthesis. In brief, 1μg total RNA containing miRNA was polyadenylated by E. coli poly (A) polymerase (NEB) at 37°C for 30 min. Then after, by using the PrimeScript™ RT reagent Kit (TaKaRa, Japan) and RT primer (Table 1), poly-adenylated RNA was reverse-transcribed. The cDNA synthesis reaction (10 μL) contained polyadenylated RNA (100 ng), 5x PrimeScript Buffer (2 μL), PrimeScript RT Enzyme Mix I (0.5 μL), RT primer mixture (1 μL equivalent 100 pmol), and RNase-free water. Then, the total reaction mixture was incubated at 50°C for 15 min and 85°C for 5 sec.
| Primers | Sequence |
|---|---|
| RT Primer | 5′- GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTTTVN-3′ (V= A, G, or C; N= A, G, C, or T.) |
| Universal Primer | 5′- GCGAGCACAGAATTAATACGACTC-3′ |
| miR-7 | 5′- GGCGTCTTTGGTTATCTAGCTGTAT-3′ |
| U6 | 5’-CGCAAGGATGACACGCAAATTC-3′ |
Table 1. Sequences of primers used in this study.
Quantitative RT-PCR for miR├ó┬?┬?7
Real-time PCR was performed with SYBR Premix Ex TaqTM II (TAKARA, Tokyo, Japan) in 15 μL final volumes with using 1.5 μL template cDNA mixed with 7.5 μL 2× SYBR Green PCR master mix and 25 pmol of each forward and reverse primers (Table 1) (Design R, 2005). We performed qPCR in duplicate according to the standard program on Rotor-Gene Q instrument (QIAGEN, Germany): 30 second at 95°C, followed by two cycles of amplification (10 sec at 95°C, 30 sec min at 60°C) and finally a dissociation curve step (ramp from 70°C to 90°C) to verify amplification specificity. Moreover, a non-template control (NTC) for each primer set included in runs to ensure reaction specificity. The change in amplification was normalized to the expression of the U6 snRNA, as internal control as it did not differ significantly between tumor and non-tumor tissue in our study population (P-value>0.05). We used the LinReg PCR software to calculate qPCR efficiencies and the fold change in expression was calculated using 2-ΔΔCt method.
Statistical analysis
Data were expressed as mean ± SEM of two or three independent experiments, and the statistical analyses were performed by PRISM 6.0 software (Graph Pad Software Inc., San Diego, CA, USA). The microRNA expression data (ΔCt values) were checked for normality using the Shapiro–Wilk test. Differences of microRNA expression amongst the groups were assessed by ordinary one-way ANOVA analysis. The Student's unpaired t-test was used to compare the microRNA expressions among two defined groups. The correlation was analyzed by Pearson correlation analysis. A p value<0.05 considered as statistically significant.
Results
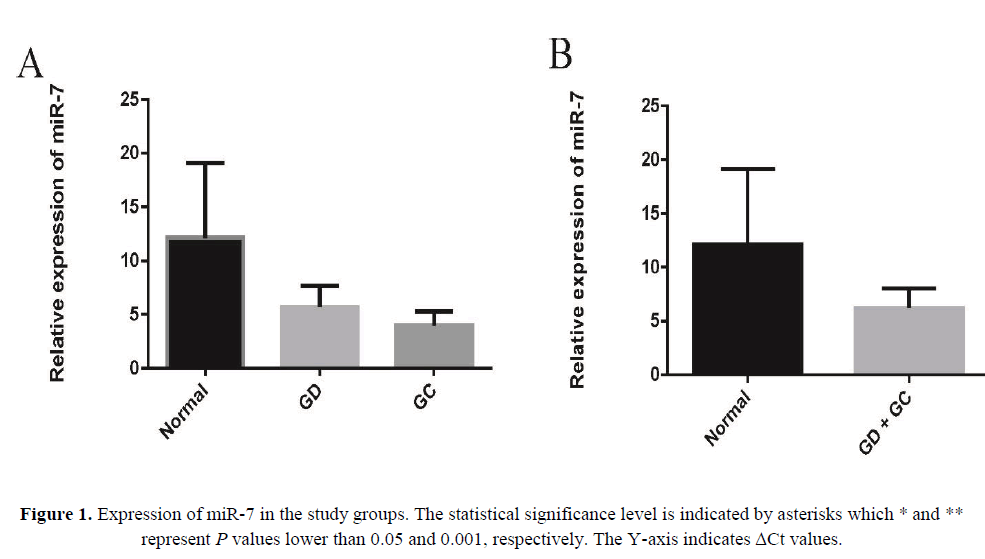
miR-7 is downregulated in GD and GC tissues
In an attempt to explore the expression and significance of miR-7 in GC carcinogenesis, we first analyzed its expression by real-time qRT– PCR in human GD and GC tissues, in which it was greatly downregulated in each compared with the control non-tumor tissues (Figure 1A). Also the results represented a more pronounced difference in microRNA expression levels when considering GC and GD sample as one group and compared to NG group (Figure 1B).
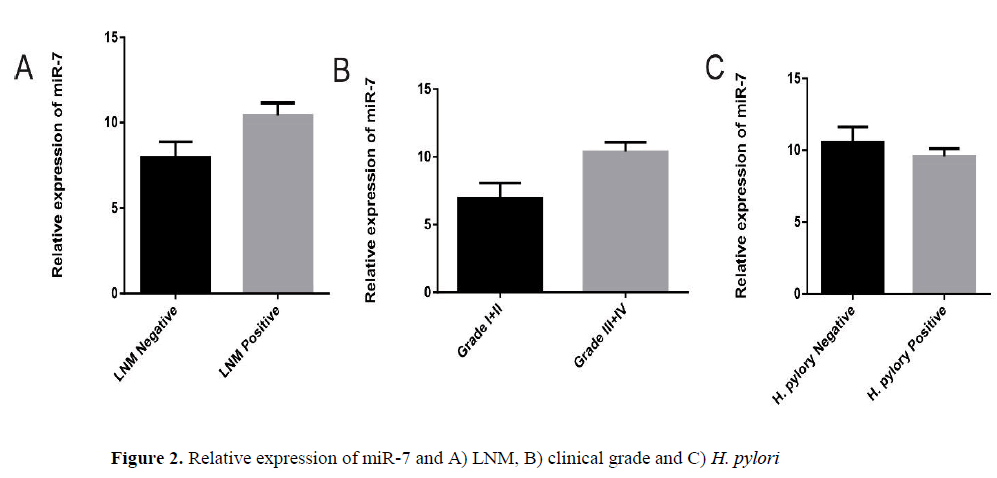
As shown in Figure 1A, the expression of miR-7 significantly reduced in GD tissues (p=0.03) and GC tissues (p=0.01) relative to their matched normal tissues. Also we found 3.48-fold decrease of miR-7 in GD and 4.92-fold decrease of miR-7 in GC tissues compared to non-tumor tissues. To check out the clinical values of miR-7, we analyzed the relationships between the expression levels of this microRNA in cancer tissues and clinico-pathological features of GC patients including age, gender, lymph node metastasis (LNM), H. pylori and clinical grade.
The expression of miR-7 is inversely correlated with LNM and higher clinical grade of gastric cancer
We analyzed the relationship between expressions of this microRNA individually, with clinicopathological factors in GC patients. A summary of the results is provided (Table 2). We observed no differences in miR-7 expression levels in GC group regarding patient’s age and sex. Our results indicated that lower expression of miR-7 is significantly correlated with lymph node metastasis (LNM) (p=0.04) and higher clinical grade (I+II vs. III+ IV, p=0.02). Compared with the GC H. pylori infected samples, the level of miR-7 expression showed no significant differences in GC samples negative for H. pylori infection (p=0.35).
| Clinico-pathological features | N | Mean | (P-value) |
|---|---|---|---|
| Age <60 |
14 | 9.733 ± 0.9765 | 0.91 |
| >60 | 26 | 9.552 ± 1.082 | |
| Sex Male |
23 | 10.51 ± 0.7968 | 0.20 |
| Female | 17 | 8.510 ± 1.418 | |
| LNM Neg |
20 | 10.39 ± 1.216 | 0.04 |
| Pos | 20 | 8.839 ± 0.9711 | |
| Grade grade I,II |
13 | 6.953 ± 1.126 | 0.01 |
| Grade III. IV | 27 | 10.38 ± 0.7061 | |
| H. pylori Neg |
14 | 10.57 ± 1.048 | 0.35 |
| Pos | 26 | 9.570 ± 0.5446 |
Table 2. The miRNAs expression in different subgroups of GC samples (n=40).
Diagnostic potential of miR-7
Since miR-7 is frequently down-regulated in gastric cancer, we tried to explore whether miR-7 expression level of gastroscopic biopsy specimen was able to predict the presence of gastric cancer. We employed Receiver Operator Characteristic (Vychytilova-Faltejskova et al., 2015) analysis and discovered that the area under the curve (AUC) was 0.849 (95% confidence interval is 0.747 to 0.952), p-value>0.0001, 83.33 sensitivity and 82.76 specificity (Figure 2).
Discussion
miRNAs have emerged as important regulators of gene expression at the post transcriptional level and regulate a wide range of physiological and developmental processes (Ambros V, 2004). Over the past several years, it has become clear that alterations in the expression of miRNAs contribute to the pathogenesis of most human cancers, where they act as either oncogenes or tumor suppressors (Lotterman CD, Kent OA, Mendell JT, 2008). Recently, accumulating data indicate that miRNAs are involved in advanced stages of cancer progression and that they can act as activators or suppressors of metastasis (Lotterman CD, Kent OA, Mendell JT, 2008; Andrews O and Patton JG, 2014).
miR-7 is downregulated in pancreatic carcinoma, non-small-cell lung cancer and cervical cancer (Bi Y, Shen W, Min M, Liu Y, 2017; Xiong S, et al., 2014; ). It has been shown that miR-7 also inhibits the motility, invasiveness, anchorage-independent growth and tumorigenic potential of highly invasive breast cancer cells (Reddy SDN, Ohshiro K, Rayala SK, Kumar R, 2008).
Our results obtained from clinical GC tissue confirm that miR-7 was downregulated in advanced stages of GC, indicating its possible involvement in both oncogenic transformation and tumor metastasis. Also, similar to classical transcription factors, miRNAs exert their effects via regulating specific target genes. Generally, one gene can be repressed by multiple miRNAs, and one miRNA may repress multiple target genes, which suggests that one specific miRNA could carry out a variety of functions by targeting different genes in different cell contexts (Price C, Chen J, 2014). Moreover, it is scientifically proven that miR-7 simultaneously in different malignancies of the digestive system (including gastric, pancreatic and colorectal cancers, and hepatocellular carcinoma) by targeting different genes and many cytokines, which indicates carcinogenic role of this non-coding RNA (Chen W-Q, Hu L, Chen G-X, Deng H-XJWjogo, 2016). Many studies have been performed to determine which genes miR-7 targets to modulate the behavior of GC metastasis (Wu X-m et al., 2011; Chen W-Q, Hu L, Chen G-X, Deng H-X, 2016). In this study, potentially target genes of miR-7 were predicted using bioinformatics databases(Imota), showed that GRB2, KRAS, EGFR, MAPK, RAF1, IGF1R, PRKCB, NRAS and FGFR4 are commonly working in Ras signaling pathway, MAPK signaling pathway and Gap junction.
Rebecca J. Webster et al. found that miR-7 down-regulates EGFR mRNA and protein expression in cancer cell lines (lung, breast, and glioblastoma) inducing cell cycle arrest and cell death (Webster RJ et al., 2009) According to another study, miR-7 expression was down-regulated in non-small cell lung cancer (NSCLC) compared with adjacent normal tissues, but the mRNA and protein expressions of FAK, ERK and MAPK were up-regulated, that can conclude the miR-7 can inhibit the activation of ERK/MAPK signaling pathway and subsequently inhibits cell proliferation, migration and invasion in human NSCLC cells (Cao Q et al., 2016). Also several studies have pointed the tumor suppressive role of miR-7 in a range of cancers (Bi Y, Shen W, Min M, Liu Y, 2017; Xiong S et al., 2014; ). Our results revealed significant down-regulation of miR-7 in pre-cancer and cancerous gastric tissues (GC vs. NG, p=0.01, NG vs. GD, p=0.03), especially in the advanced grades and LNM episodes, highlighting its roles in tumor development and progression.
Also, there are evidence suggested that miR-7 expressed lower levels LNM positive tissues compared with primary GC tissues and the adjacent normal tissue, indicating an inverse relationship between the expression of miR-7 and the metastatic status of GC tissues (Zhao X et al., 2013). Moreover, Zhao et al. showed that miR-7/IGF1R/Snail axis are involved in pathogenesis of GC. They found that miR-7 is inversely correlated with IGF1R expression which indicates the anti-metastatic role of miR-7 (Zhao X et al., 2013). Supporting evidence suggested that overexpression of insulin-like growth factor-1 receptor mediate cancer cell metastasis and has been reported in different cancers (Zhao X et al., 2013). Such findings suggest microRNA-7 might play a tumor suppressive role through regulating Ras signaling pathway, MAPK signaling pathway and Gap junction. Emerging evidence describe the modulation of mammalian miRNAs in response to bacterial infection (Eulalio A, Schulte L, Vogel J, 2012). H. pylori infection has been associated with alterations in the miRNA expression profile in gastric tissue and known to be a major risk factor for GC (Noto JM and Peek Jr RM, 2012). Although regarding miR-7 expression, no statistically significance was observed.
In recent years, identification and characterization of premalignant lesions including GD proposed to help to stratify the individual risk for GC development (Marqués-Lespier JM, González-Pons M, Cruz-Correa M, 2016 ). The current pathology-based systems of gastric cancer have failed to efficiently predict the risk of sequential development from premalignant lesions such as GD to malignant lesions. The molecular signatures of these transitions have not been investigated widely; however, some genetic alterations have been proposed to be involved (Yin Y et al., 2012). As Figure 1A and 1B represent, expression levels of miR-7 were significantly different when NG group was compared to GD alone or together with GC samples. Such results indicate that down-regulation of this miRNAs might be served as a signature of sequential transition from NG to GD and eventually GC.
Conclusion
In conclusion we found down-regulation of miR-7 in pre-cancerous and cancerous tissues. Our findings further implied that this down-regulated miRNA is markedly changed biomarker in GD and GC as compared to NG. In this regard new light should be shed on this miRNA role in neoplastic transition. However, one must keep in mind that the findings in our study were based on a limited number of samples and clinicopathological parameters, and require confirmation from the analysis of larger cohorts.
DECLARATIONS
Ethics approval and consent to participants
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Consent of publication
Not applicable.
Availability of data and materials
The analysed data sets generated during the study are available from the corresponding author on reasonable request.
Competing Interest
The authors declare they have no conflict of interest.
Funding
Not applicable.
Authors' Contributions
Mir Davood Omrani and Hamid Ghaedi supervised the study. Sara Omrani and Ali Zare done the laboratory assessment. Shahriar Tarighi Analysed the data.
Acknowledgment
The current study was supported by a grant from Shahid Beheshti University of Medical Sciences.
About the Authors
Corresponding Author
Mir Davood Omrani
Faculty of Medicine, Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- Email:
- infolib@sbmu.ac.ir
References
- Tan YK and Fielding JW (2006) Early diagnosis of early gastric cancer. Eur J Gastroenterol Hepatol 18: 821-829. https://doi.org/10.1097/00042737-200608000-00004
- Fitzmaurice C, Dicker D, Pain A, Hamavid H, et al. (2015) The global burden of cancer 2013. JAMA oncology 1:505-527.
- Yonemura Y, Fujimura T, Fushida S, Takegawa S, et al. (1991) Hyperthermo-chemotherapy combined with cytoreductive surgery for the treatment of gastric cancer with peritoneal dissemination. World J Surg 15: 530-535. https://doi.org/10.1007/bf01675656
- Ryu KW, Mok YJ, Kim SJ, Kim CS (2000) Prognostic factors in advanced gastric cancer with peritoneal carcinomatosis. J Korean Surg Soc 59: 786-792.
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350-355. https://doi.org/10.1038/nature02871
- Jay C, Nemunaitis J, Chen P, Fulgham P, et al. (2007) miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol 26: 293-300. https://doi.org/10.1089/dna.2006.0554
- Lotterman CD, Kent OA, Mendell JT (2008) Functional integration of microRNAs into oncogenic and tumor suppressor pathways. Cell Cycle 7: 2493-2499. https://doi.org/10.4161/cc.7.16.6452
- Gao W, Yu Y, Cao H, Shen H, et al. (2010) Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed Pharmacother 64: 399-408. https://doi.org/10.1016/j.biopha.2010.01.018
- Vychytilova-Faltejskova P, Kiss I, Klusova S, Hlavsa J, et al. (2015) MiR-21, miR-34a, miR-198 and miR-217 as diagnostic and prognostic biomarkers for chronic pancreatitis and pancreatic ductal adenocarcinoma. Diagn Pathol 10: 38. https://doi.org/10.1186/s13000-015-0272-6
- Wang F, Zheng Z, Guo J, Ding X (2010) Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol 119: 586-593. https://doi.org/10.1016/j.ygyno.2010.07.021
- Guo J, Miao Y, Xiao B, Huan R, et al. (2009) Differential expression of microRNA species in human gastric cancer versus non├ó┬?┬Étumorous tissues. J Gastroenterol Hepatol 24: 652-657. https://doi.org/10.1111/j.1440-1746.2008.05666.x
- Horsham JL, Kalinowski FC, Epis MR, Ganda C, et al. (2015) Clinical potential of microRNA-7 in cancer. J Clin Med 4: 1668-1687. https://doi.org/10.3390/jcm4091668
- Bi Y, Shen W, Min M, Liu Y (2017) MicroRNA-7 functions as a tumor-suppressor gene by regulating ILF2 in pancreatic carcinoma. Int J Mol Med 39: 900-906. https://doi.org/10.3892/ijmm.2017.2894
- Xiong S, Zheng Y, Jiang P, Liu R, et al. (2014) PA28gamma emerges as a novel functional target of tumour suppressor microRNA-7 in non-small-cell lung cancer. Br J Cancer 110: 353-362. https://doi.org/10.1038/bjc.2013.728
- Hao Z, Yang J, Wang C, Li Y, et al. (2015 ) MicroRNA-7 inhibits metastasis and invasion through targeting focal adhesion kinase in cervical cancer. Int J Clin Exp Med 8: 480.
- Design R (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519-525. https://doi.org/10.2144/000112010
- Andrews O and Patton JG (2014) MicroRNAs in cancer progression. MicroRNA in Development and in the Progression of Cancer: Springer p. 29-46. https://doi.org/10.1007/978-1-4899-8065-6_2
- Reddy SDN, Ohshiro K, Rayala SK, Kumar R (2008) MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer res 68: 8195-8200. https://doi.org/10.1158/0008-5472.can-08-2103
- Price C and Chen J (2014) MicroRNAs in cancer biology and therapy: Current status and perspectives. Genes & diseases 1: 53-63. https://doi.org/10.1016/j.gendis.2014.06.004
- Chen W-Q, Hu L, Chen G-X, Deng H-XJWjogo (2016) Role of microRNA-7 in digestive system malignancy 8: 121. https://doi.org/10.4251/wjgo.v8.i1.121
- Wu X-m, Shao X-q, Meng X-x, Zhang X-n, et al. (2011) Genome-wide analysis of microRNA and mRNA expression signatures in hydroxycamptothecin-resistant gastric cancer cells. Acta Pharmacologica Sinica 32: 259. https://doi.org/10.1038/aps.2010.204
- Chen W-Q, Hu L, Chen G-X, Deng H-X (2016) Role of microRNA-7 in digestive system malignancy. World J Gastrointest Oncol 8: 121. https://doi.org/10.4251/wjgo.v8.i1.121
- Webster RJ, Giles KM, Price KJ, Zhang PM, et al. (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 284: 5731-5741. https://doi.org/10.1074/jbc.m804280200
- Cao Q, Mao ZD, Shi YJ, Chen Y, et al. (2016) MicroRNA-7 inhibits cell proliferation, migration and invasion in human non-small cell lung cancer cells by targeting FAK through ERK/MAPK signaling pathway. Oncotarget 7: 77468. https://doi.org/10.18632/oncotarget.12684
- Zhao X, Dou W, He L, Liang S, et al. (2013) MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor 32: 1363. https://doi.org/10.1038/onc.2012.156
- Zhao X, Dou W, He L, Liang S, et al. (2013) MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 32: 1363-1372. https://doi.org/10.1038/onc.2012.156
- Eulalio A, Schulte L, Vogel J (2012) The mammalian microRNA response to bacterial infections. RNA Biol 9: 742-750. https://doi.org/10.4161/rna.20018
- Noto JM and Peek Jr RM (2012) The role of microRNAs in Helicobacter pylori pathogenesis and gastric carcinogenesis. Front Cell Infect Microbiol 1: 21. https://doi.org/10.3389/fcimb.2011.00021
- Marqués-Lespier JM, González-Pons M, Cruz-Correa M (2016 ) Current perspectives on gastric cancer. Gastroenterol Clin 45: 413-428. https://doi.org/10.1016/j.gtc.2016.04.002
- Yin Y, Li J, Chen S, Zhou T, et al. (2012) MicroRNAs as diagnostic biomarkers in gastric cancer. Int J Mol Sci 13: 12544-12555. https://doi.org/10.3390/ijms131012544
Keywords:
Download:
Full PDF- Share This