Relationship between melanoma-associated antigen 1 (MAGE-A1) gene polymorphisms and colorectal cancer development
Received: October 25, 2018
Accepted: November 12, 2018
Published: January 05, 2019
Genet.Mol.Res. 18(4):
Introduction
Colorectal Cancer (CRC) is a heterogeneous disease; it occurs through combinations of different factors, such as environments, genetics and epigenetics (Kruk J and Czerniak U, 2013; Peters U et al., 2013). In the United States, it is the second-most major neoplasm and leads to death among both males and females. Approximately 140,000 Americans are diagnosed with this disease and more than 50,000 related deaths are recorded annually (Al-Thafar AK et al., 2017). In the Kingdom of Saudi Arabia (KSA), this malignancy affects both men and women and is the second-leading cause of cancer-related deaths in KSA citizens (Saudi Cancer Registry, 2011). Moreover, it is the first- and third-most common cancer among Saudi men and women, respectively (Al-Thafar AK et al., 2017). Additionally, CRC mortalities and incidences in the KSA have recently increased, leading to higher risks of increased CRC mortality rates in the KSA (Ibrahim EM et al., 2008). To improve these tests and decrease CRC deaths, several studies investigated genes associated with CRC regarding prognoses and screening-related biomarkers (Krupa R et al., 2011; Lurje G, Zhang W, Lenz HJ 2007; Peltekova VD et al., 2014) However, early CRC biomarkers that could help predict the presence of the disease were not identified. Despite this, the analysis of genetic variations may provide such an insight into the genetics of human CRC. For example, SNPs, the most common genetic variations in the human genome, may be excellent genotypic biomarkers for predicting CRC developments as they have the potential to be unique genetic markers for association-based approaches to discovering genetic components (Ulrich CM, Robien K, McLeod HL, 2003).
In addition, SNP patterns and profiles may assist in identifying comprehensive gene collections that may contribute to cancer developments and susceptibility (Semlali A et al., 2016). The presence of SNPs in several genes, such as CYP24-A1 (Chen XQ et al., 2017), TLR-9 (Semlali A et al., 2016). RETN (Alharithy RN, 2014), and TNFα (Kapitanovic S et al., 2014), was found to be associated with colon cancer risks. Moreover, SNPs were found to be located in different positions within genes, such as in promoters, exons, introns, and 5' or, 3' untranslated regions (UTRs) and these SNPs can alter gene expressions using various mechanisms (Deng N, Zhou H, Fan H, Yuan Y, 2017). Therefore, this study aimed to investigate the potential contribution of melanoma-associated antigen 1 (MAGE-A1) SNPs (rs3788753, rs3788749 and rs3788745) and expression levels in CRC among Saudi patients compared with healthy colorectal samples to identify a diagnostic biomarker for patients with CRC.
It should be noted that cancerous human antigens can generally be separated into five categories: cancer/testis (CT) antigens, overexpressed proteins, tumor viruses, mutated antigens and differentiation antigens (Stevanovic S, 2002). CT genes are a large category of testis-specific genes that are expressed in human cancers of diverse histological origins (Babatunde KA et al., 2017; Scanlan MJ, Simpson AJ, Old LJ, 2004). These include melanomas, ovarian cancers, colon cancers, pancreatic cancers, lung malignancies, renal malignancies and hematopoietic malignancies (Caballero OL and Chen YT, 2009). Among the CTAs identified to date, the MAGE family is one of the most widely studied antigen families that recognize expressions in a wide variety of human tumors (Simpson AJG et al., 2005). “MAGE” was originally a “melanoma-associated antigen”; later it was discovered that its homologs formed a multi-gene family in mammalian genomes (Vanderbruggen P et al., 1991; Castelli C et al., 2000).
In the human genome, this family can be categorized into three subfamilies based on expression patterns and functions (Vanderbruggen P et al., 1991; Katsura Y and Satta Y, 2011): there are 12 MAGEA, 18 MAGEB and 7 MAGEC (Doyle JM, Gao J, Wang J, Yang M, 2010; Chomez P et al., 2001); members. Each subfamily comprises 1–15 genes [21, 23], and is expressed in highly proliferating cells, such as tumor, placenta, and germ line cells (Vanderbruggen P et al., 1991; Katsura Y and Satta Y, 2011). MAGEA, MAGEB and MAGEC genes are located on the X chromosome, and they encode tumor antigens that are important in cancer immunity. Peptides in the human MAGE homology domain (MHD) are 160–170 amino acids long (Vanderbruggen P et al., 1991; Katsura Y and Satta Y, 2011). Melanoma Associated Antigen-A1 (MAGE-A1) was identified as the first CTA in a melanoma patient (Vanderbruggen P et al., 1991).
Incidences of CRC in the Saudi population have increased in recent years. Consequently, screening processes are important for the early detection of CRC and decreasing the CRC death rate. Therefore, this study investigated the possibility of using MAGE-A1common primer sets to screen for CRC among Saudi patients. However, while MAGE-A1had many SNPs, there was no evidence these SNPs could be used as tumor prognosis factors among patients with CRC. To confirm the role of MAGE-A1polymorphisms in tumor prognoses of patients with CRC, three MAGE-A1SNPs (rs3788753, rs3788749 and rs3788745) were analyzed. Moreover, the allele and genotype frequencies of MAGE-A1polymorphisms and clinical factors, such as age, sex, and pathological classifications were analyzed among Saudi patients with CRC compared to matched normal colorectal tissues in order to determine potential influences on CRC susceptibility.
Material and Methods
Ethical approval certificate and data collection
This case-controlled study was reviewed by, and ethical approval was obtained from, the Ethics Review Committee of the Medicine College at the King Khalid University Hospital (KKUH) of King Saud University, Riyadh, Saudi Arabia. Written informed consent was obtained from participants after the objectives of this study were explained. Each participating subject completed and signed an informed consent questionnaire regarding his or her participation before blood sample was taken for genotyping purposes and a tissue biopsy was conducted for gene expression purposes. No patients received either immunotherapy or radiotherapy prior to sample collection, and the questionnaire collected demographic and clinical data, such as the age at which each patient was diagnosed with CRC, as well as each patient’s gender, family health history, smoking habits, tumor location, tumor grade and date of diagnosis. The questionnaire was written in both Arabic and English to ensure participants found it easy to understand, and the patients agreed that all data could be used for publishing.
Population and sample collection
To study MAGE-A1polymorphisms, the participants were divided into two groups. The first was a CRC group that comprised 192 Saudi patients (114 men and 78 women) with histological diagnoses of CRC and positive clinical, endoscopic, radiological and histopathological results for malignancies between 2011 and 2016 through the endoscopy service of the KKUH before they received any medical treatments for the disease. The second was a control group that comprised 192 age-matched healthy individuals (111 men and 81 women) who lacked any signs of malignancy. Whole blood specimens were collected from the individuals and genomic DNA was extracted from the blood to study the MAGE-A1polymorphisms of the CRC causes in comparison with normal groups.
DNA extraction from peripheral blood
A total of 3 ml of blood was taken from each study participant using an EDTA Vacutainer vial (BD Vacutainer Systems, Plymouth, UK), then stored at −80°C until required. Genomic DNA was immediately extracted from 200 μl of EDTA-anticoagulated whole blood in accordance with the manufacturer’s instructions using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA). The isolated genomic DNA was preserved at −20°C until analysis, and the concentration of the DNA samples was validated via a NanoDrop 8000 spectrophotometer (Thermo Scientific, USA). Finally, DNA purity was measured in terms of A260/A280 and A260/A230 absorbance ratios.
MAGE SNP selections and DNA genotyping
TaqMan Allelic Discrimination assay kits (Thermo Fisher Scientific, Waltham, MA, USA) were used to genotype the three MAGE-A1polymorphisms: rs3788753 (C/G), rs3788749 (G/A), and rs3788745 (A/G). All primer sequences and probe mixtures that were used were designed by Thermo Fisher Scientific as per the manufacturer’s protocol. MAGE-A1polymorphisms were selected based on three different criteria: MAGE-A1SNP positions, MAGE-A1SNP frequencies with cancer issues, and MAGE-A1SNP correlations with different malignances among patients of different ethnic backgrounds. For DNA genotyping, each sample was examined using a 96-Well Format QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Briefly, reaction mixtures were briefly investigated using a 10μL/reaction volume. Each reaction contained 2 μL of DNA (20 ng), 0.2 μL of 40× TaqMan® Genotyping SNP Assay (Applied Biosystems), 5.5 μL of 2X TaqMan PCR Master Mix (Applied Biosystems) and 2.3 μL of distilled water. For PCR amplifications, a Light Cycler 480 Instrument II Real Time PCR System was used.
The efficiency of genotyping samples was tested to confirm the genotyping quality with positive and negative controls. The laboratory staff that performed the genotyping assays was blinded to patient’s information.
The statistical methods used for data analyses
Data analyses were conducted using the Statistical Package for Social Sciences (SPSS, version 24, made by SPSS, Chicago, USA). p-values were considered statistically significant is they were below 0.05, and analyzed polymorphism frequencies were tested for patients with CRC and healthy controls using Hardy-Weinberg equilibrium (HWE). Odds ratios (ORs) and 95% confidence intervals (CIs) were adjusted for both genders and ages. Χ2 values were measured using chi-square tests to determine significant differences between the patients with CRC and the healthy individuals.
Results
The basic clinical parameters of the study populations
As shown in Table 1, 192 cases with pathologically confirmed CRC and 192 normal volunteers were enrolled, and any patient who was not Saudi and/or who was a smoker was excluded from the study. There were no significant demographic variations between the CRC cases and the healthy controls; however, there were some clinical differences. For example, patients’ ages, genders and tumor locations varied between groups. The median ages were 55.74 ± 12.34 years and 56.56 ± 11.21 years for healthy and cancer groups, respectively. Moreover, no significant differences were found in the male-to-female ratios of the CRC cases and the normal volunteers (the ratios were 114:78 and 111:81 for CRC and control individuals, respectively). Other clinical parameters for the populations, including the localizations of the patients’ tumors and CRC therapy data, are shown in Table 1.
| Variables | Cancer | Control |
|---|---|---|
| Number | 192 | 192 |
| Gender | ||
| Male | 114 | 111 |
| Female | 78 | 81 |
| Age | ||
| Below 57 | 106 | 94 |
| Above 57 | 86 | 98 |
| Median Age | 56.56± 11.21 | 55.74 ± 12.34 |
| Localization | ||
| Colon | 116 | --- |
| Rectum | 76 | --- |
Table 1: Clinical characteristics of the subjects for genotyping.
The relationship between select MAGE-A1polymorphisms and CRC risks among Saudi patients
As previously mentioned, to examine the correlation between MAGE-A1polymorphisms and colorectal cancer susceptibility among the Saudi population, three MAGE-A1SNPs (rs3788753, rs3788749 and rs3788745) were examined and their allelic and genotypic frequencies were compared between 192 CRC patients and 192 healthy control individuals. The allelic and genotype distributions of the examined SNPs, along with corresponding odds ratios, are shown in Table 3. Observations of genotypic frequencies did not result in any significant Hardy–Weinberg expectations for rs3788745 and rs3788753. The genotype frequencies of patients with CRC were 91% for TT, 8% for TC and 1% for CC for rs3788745 and 90% CC, 5% CG and 5% GG for rs3788753. The genotype frequencies of control individuals were 92%, 6% and 2% for TT, TC and CC for rs3788745, respectively, and 88%, 8% and 4% for CC, CG and GG for rs3788753, respectively. However, the heterozygous CT and a combination of CT + TT genotypes for the SNP rs3788749 showed a more than two-fold higher risk for CRC developments in patients with CRC, not the healthy control individuals. For CT, OR was 2.528, the CI was 1.023–6.248 and p= 0.038. For CT + TT, OR was 2.677, the CI was 1.091–6.570 and p= 0.026. Moreover, the T allele frequency was high in CRC cases, unlike the control cases; OR was 2.745, the CI was 1.140–6.609 and p= 0.019 (Table 2).
| SNP | Variants | Patients Cases | Controls | OR | CI | χ2 Value | P- Value |
|---|---|---|---|---|---|---|---|
| rs3788745 | TT | 172 (0.91) | 175 (0.92) | Ref | |||
| TC | 16 (0.08) | 11 (0.06) | 1.480 | 0.668-3.280 | 0.94 | 0.33197 | |
| CC | 2 (0.01) | 4 (0.02) | 0.509 | 0.092-2.814 | 0.62 | 0.43034 | |
| TC+CC | 18 (0.9) | 15 (0.08) | 1.221 | 0.596-2.500 | 0.30 | 0.58472 | |
| T | 360 (0.95) | 361 (0.95) | Ref | ||||
| C | 20 (0.05) | 19 (0.05) | 1.056 | 0.554-2.011 | 0.03 | 0.86941 | |
| rs3788749 | CC | 171 (0.90) | 178 (0.96) | Ref | |||
| CT | 17 (0.09) | 7 (0.04) | 2.528 | 1.023-6.248 | 4.28 | 0.03849 | |
| TT | 1 (0.01) | 0 (0.00) | 3.122 | 0.126-77.175 | 1.04 | 0.30832 | |
| CT+TT | 18 (0.10) | 7 (0.04) | 2.677 | 1.091-6.570 | 4.94 | 0.02627 | |
| C | 359 (0.95) | 363 (0.98) | Ref | ||||
| T | 19 (0.05) | 7 (0.02) | 2.745 | 1.140-6.609 | 5.48 | 0.01928 | |
| rs3788753 | CC | 173 (0.90) | 166 (0.88) | Ref | |||
| CG | 10 (0.05) | 14 (0.08) | 0.685 | 0.296-1.586 | 0.79 | 0.37517 | |
| GG | 9 (0.05) | 8 (0.04) | 1.079 | 0.407-2.864 | 0.02 | 0.87790 | |
| CG+GG | 19 (0.10) | 22 (0.12) | 0.829 | 0.433-1.587 | 0.32 | 0.57042 | |
| C | 356 (0.93) | 346 (0.92) | Ref | ||||
| G | 28 (0.07) | 30 (0.08) | 0.907 | 0.531-1.550 | 0.13 | 0.72133 |
Notes: P-Values in bold represent significant results (P < 0.05); Ref=Reference allele.
Table 2: Genotype frequencies of MAGEA1 gene polymorphism in colorectal cases and controls.
| Gender | SNP | Variant | patients Cases | Controls | OR | CI | χ2 Value | P- Value |
|---|---|---|---|---|---|---|---|---|
| Male | rs3788745 | TT | 104 (0.92) | 101 (0.93) | Ref | |||
| TC | 8 (0.07) | 5 (0.05) | 2.200 | 0.738-6.560 | 2.09 | 0.14861 | ||
| CC | 1 (0.01) | 3 (0.02) | 0.333 | 0.034-3.259 | 0.98 | 0.32203 | ||
| TC+CC | 9 (0.08) | 8 (0.07) | 1.500 | 0.588-3.825 | 0.73 | 0.39348 | ||
| T | 216 (0.96) | 207 (0.95) | Ref | |||||
| C | 10 (0.04) | 11 (0.05) | 1.149 | 0.503-2.622 | 0.11 | 0.74212 | ||
| rs3788749 | CC | 101 (0.89) | 104 (0.95) | Ref | ||||
| CT | 11 (0.10) | 5 (0.05) 0.) |
2.265 | 0.760-6.751 | 2.25 | 0.13330 | ||
| TT | 1 (0.01) | 0 (0.00) | 3.089 | 0.124-76.706 | 1.02 | 0.31143 | ||
| CT+TT | 12 (0.11) | 5 (0.05) | 2.471 | 0.840-7.267 | 2.86 | 0.09108 | ||
| C | 213 (0.94) | 213 (0.98) | Ref | |||||
| T | 13 (0.06) | 5 (0.02) | 2.600 | 0.911-7.421 | 3.41 | 0.06470 | ||
| rs3788753 | CC | 104 (0.91) | 96 (0.88) | Ref | ||||
| CG | 6 (0.05) | 6 (0.06) | 0.923 | 0.288-2.960 | 0.02 | 0.89286 | ||
| GG | 4 (0.04) | 6 (0.06) | 0.615 | 0.169-2.247 | 0.55 | 0.45872 | ||
| CG+GG | 10 (0.09) | 12 (0.12) | 0.769 | 0.318-1.862 | 0.34 | 0.55988 | ||
| C | 214 (0.94) | 198 (0.92) | Ref | |||||
| G | 14 (0.06) | 18 (0.08) | 0.720 | 0.349-1.485 | 0.80 | 0.37181 | ||
| Female | rs3788745 | TT | 68 (0.88) | 73 (0.91) | Ref | |||
| TC | 8 (0.10) | 6 (0.08) | 1.043 | 0.321-3.388 | 0.0002 | 0.94434 | ||
| CC | 1 (0.01) | 1 (0.01) 0.01) |
0.348 | 0.014-8.674 | 0.95 | 0.32907 | ||
| TC+CC | 9 (0.12) | 7 (0.09) | 0.894 | 0.286-2.791 | 0.04 | 0.84681 | ||
| T | 144 (0.94) | 152 (0.95) | Ref | |||||
| C | 10 (0.06) | 8 (0.05) | 0.781 | 0.264-2.305 | 0.20 | 0.65349 | ||
| rs3788749 | CC | 70 (0.92) | 73 (0.97) | Ref | ||||
| CT | 6 (0.08) | 2 (0.03) | 3.129 | 0.611-16.025 | 2.06 | 0.15157 | ||
| TT | 0 (0.00) | 0 (0.00) | ||||||
| CT+TT | 6 (0.08) | 2 (0.03) | 3.129 | 0.611-16.025 | 2.06 | 0.15157 | ||
| C | 146 (0.96) | 148 (0.97) | Ref | |||||
| T | 6 (0.04) | 2 (0.03) | 3.041 | 0.604-15.314 | 2.00 | 0.28294 | ||
| rs3788753 | CC | 69 (0.88) | 70 (0.87) | Ref | ||||
| CG | 4 (0.06) | 8 (0.10) | 0.507 | 0.146-1.762 | 1.18 | 0.27812 | ||
| GG | 5 (0.06) | 2 (0.03) | 2.536 | 0.476-13.516 | 1.27 | 0.26057 | ||
| CG+GG | 9 (0.12) | 10 (0.13) | 0.913 | 0.350-2.385 | 0.03 | 0.85262 | ||
| C | 142 (0.91) | 148 (0.92) | Ref | |||||
| G | 14 (0.09) | 12 (0.08) | 1.216 | 0.544-2.719 | 0.23 | 0.63347 |
Ref=Reference allele
Table 3: Genotype frequencies of MAGEA1 gene polymorphism in colorectal cases and controls (gender: Male or Female)
Comparison of MAGE-A1SNP genotype and allele allocations based on the clinical parameters of the CRC and control subjects
MAGE-A1 genotypic and allelic distributions were analyzed after correlations between genders, ages, and tumor locations and CRC diagnoses were determined. First, the relationship between CRC risks and individual SNPs was determined based on genders. The genotype frequencies of males (n=114) and females (n=78) with CRC, compared to those of healthy males and females, are presented in Table 4. No significant relationship was found between female and male patients with CRC and healthy male and female subjects (see Table 3 for males and females). Second, SNP correlations to the ages at which patients were diagnosed with CRC were analyzed and patients were categorized as ≤ 57 (n=106) or > 57 (n=86) years of age. The allele and genotype allocations of individual SNPs and statistical analyses are shown in Table 5 and 6, respectively.
| Age | SNP | Variant | patients Cases | Controls | OR | CI | χ2 Value | P- Value |
|---|---|---|---|---|---|---|---|---|
| Below 57 | rs3788745 | TT | 95 (0.91) | 84 (0.92) | Ref | |||
| TC | 9 (0.09) | 5 (0.05) | 1.987 | 0.663-5.955 | 1.55 | 0.21296 | ||
| CC | 0 | 2 (0.03) | 0.181 | 0.009-3.819 | 2.19 | 0.13916 | ||
| TC+CC | 9 (0.09) | 7 (0.08) | 1.419 | 0.526-3.829 | 0.48 | 0.48752 | ||
| T | 199 (0.96) | 173 (0.95) | Ref | |||||
| C | 9 (0.04) | 9 (0.05) | 1.073 | 0.435-2.651 | 0.02 | 0.87809 | ||
| rs3788749 | CC | 93 (0.89) | 85 (0.94) | Ref | ||||
| CT | 11 (0.11) | 5 (0.06) | 2.011 | 0.671-6.024 | 1.61 | 0.20483 | ||
| TT | 0 (0.00) | 0 (0.00) | ||||||
| CT+TT | 11 (0.11) | 5 (0.06) | 2.011 | 0.671-6.024 | 1.61 | 0.20483 | ||
| C | 197 (0.95) | 175 (0.97) | Ref | |||||
| T | 11 (0.05) | 5 (0.03) | 1.954 | 0.666-5.735 | 1.54 | 0.21485 | ||
| rs3788753 | CC | 96 (0.91) | 77 (0.85) | Ref | ||||
| CG | 4 (0.04) | 8 (0.09) | 0.401 | 0.116-1.382 | 2.22 | 0.13637 | ||
| GG | 5 (0.05) | 5 (0.06) 0) |
0.802 | 0.224-2.871 | 0.12 | 0.73422 | ||
| CG+GG | 9 (0.09) | 13 (0.15) | 0.555 | 0.225-1.367 | 1.67 | 0.19626 | ||
| C | 196 (0.93) | 162 (0.90) | Ref | |||||
| G | 14 (0.07) | 18 (0.10) | 0.643 | 0.310-1.332 | 1.43 | 0.23179 | ||
| Above 57 | rs3788745 | TT | 76 (0.89) | 90 (0.92) | Ref | |||
| TC | 7 (0.08) | 6 (0.06) | 1.169 | 0.362-3.773 | 0.07 | 0.79397 | ||
| CC | 2 (0.02) | 2 (0.02) | 0.584 | 0.052-6.570 | 0.19 | 0.65987 | ||
| TC+CC | 9 (0.11) | 8 (0.08) | 1.023 | 0.355-2.949 | 0.004 | 0.96682 | ||
| T | 159 (0.94) | 186 (0.95) | Ref | |||||
| C | 11 (0.06) | 10 (0.05) | 0.930 | 0.358-2.413 | 0.02 | 0.88138 | ||
| rs3788749 | CC | 77 (0.92) | 92 (0.98) | Ref | ||||
| CT | 6 (0.07) | 2 (0.02) | 3.584 | 0.703-18.270 | 2.66 | 0.10303 | ||
| TT | 1 (0.01) | 0 (0.00) | ||||||
| CT+TT | 7 (0.08) | 2 (0.02) | 4.182 | 0.844-20.720 | 3.56 | 0.05924 | ||
| C | 160 (0.95) | 186 (0.99) | Ref | |||||
| T | 8 (0.05) | 2 (0.01) | 4.650 | 0.973-22.214 | 4.44 | 0.05128 | ||
| rs3788753 | CC | 76 (0.88) | 89 (0.91) | Ref | ||||
| CG | 6 (0.07) | 6 (0.06) | 1.171 | 0.363-3.782 | 0.07 | 0.79160 | ||
| GG | 4 (0.05) | 3(0.03) | 1.561 | 0.339-7.196 | 0.33 | 0.56477 | ||
| CG+GG | 10 (0.12) | 9 (0.09) | 1.301 | 0.503-3.368 | 0.30 | 0.58670 | ||
| C | 158 (0.92) | 184 (0.94) | Ref | |||||
| G | 14 (0.08) | 12 (0.06) | 1.359 | 0.611-3.023 | 0.57 | 0.45119 |
Ref=Reference allele
Table 4: Genotype frequencies of MAGEA1 gene polymorphism in colorectal cases and controls (age: below 57 or above 57)
| Tumor location | SNP | Variant | patients Cases | Controls | OR | CI | χ2 Value | P- Value |
|---|---|---|---|---|---|---|---|---|
| Colon | rs3788745 | TT | 101 (0.89) | 175 (0.92) | Ref | |||
| TC | 10 (0.09) | 11 (0.06) | 1.575 | 0.646-3.838 | 1.01 | 0.31408 | ||
| CC | 2 (0.02) | 4 (0.02) | 0.866 | 0.156-4.814 | 0.03 | 0.86964 | ||
| TC+CC | 12 (0.11) | 15 (0.08) | 1.386 | 0.624-3.078 | 0.65 | 0.42079 | ||
| T | 212 (0.94) | 361 (0.95) | Ref | |||||
| C | 14 (0.06) | 19 (0.05) | 1.255 | 0.616-2.554 | 0.39 | 0.53081 | ||
| rs3788749 | CC | 103 (0.92) | 178 (0.96) | Ref | ||||
| CT | 8 (0.07) | 7 (0.04) | 1.975 | 0.696-5.605 | 1.69 | 0.19359 | ||
| TT | 1 (0.01) | 0 (0.00) | 5.174 | 0.209-128.173 | 1.72 | 0.19000 | ||
| CT+TT | 9 (0.08) | 7 (0.04) | 2.222 | 0.804-6.144 | 2.47 | 0.11570 | ||
| C | 214 (0.96) | 363 (0.98) | Ref | |||||
| T | 10 (0.04) | 7 (0.02) | 2.423 | 0.909-6.460 | 3.32 | 0.06839 | ||
| rs3788753 | CC | 102 (0.89) | 166 (0.88) | Ref | ||||
| CG | 6 (0.05) | 14 (0.08) | 0.697 | 0.260-1.873 | 0.52 | 0.47263 | ||
| GG | 7 (0.06) | 8 (0.04) | 1.424 | 0.501-4.045 | 0.44 | 0.50502 | ||
| CG+GG | 13 (0.11) | 22 (0.12) | 0.962 | 0.464-1.993 | 0.01 | 0.91628 | ||
| C | 210 (0.91) | 346 (0.92) | Ref | |||||
| G | 20 (0.09) | 30 (0.08) | 1.098 | 0.608-1.984 | 0.10 | 0.75559 | ||
| Rectum | rs3788745 | TT | 70 (0.92) | 175 (0.92) | Ref | |||
| TC | 6 (0.08) | 11 (0.06) | 1.364 | 0.486-3.830 | 0.35 | 0.55475 | ||
| CC | 0 | 4 (0.02) | 0.277 | 0.015-5.205 | 1.59 | 0.20736 | ||
| TC+CC | 6 (0.08) | 15 (0.08) | 1.000 | 0.373-2.682 | 0.001 | 1 | ||
| T | 146 (0.96) | 361 (0.95) | Ref | |||||
| C | 6 (0.04) | 19 (0.05) | 0.781 | 0.306-1.994 | 0.27 | 0.60426 | ||
| rs3788749 | CC | 67 (0.88) | 178 (0.96) | Ref | ||||
| CT | 9 (0.12) | 7 (0.04) | 3.416 | 1.223-9.538 | 6.08 | 0.01368 | ||
| TT | 0 (0.00) | 0 (0.00) | 2.644 | 0.052-134.610 | 0.001 | 1 | ||
| CT+TT | 9 (0.12) | 7 (0.04) | 3.416 | 1.223-9.538 | 6.08 | 0.01368 | ||
| C | 143 (0.94) | 363 (0.98) | Ref | |||||
| T | 9 (0.06) | 7 (0.02) | 3.264 | 1.193-8.930 | 5.89 | 0.02373 | ||
| rs3788753 | CC | 70 (0.92) | 166 (0.88) | Ref | ||||
| CG | 4 (0.05) | 14 (0.08) | 0.678 | 0.215-2.131 | 0.45 | 0.50317 | ||
| GG | 2 (0.03) | 8 (0.04) | 0.593 | 0.123-2.862 | 0.43 | 0.51075 | ||
| CG+GG | 6 (0.08) | 22 (0.12) | 0.647 | 0.251-1.664 | 0.83 | 0.36300 | ||
| C | 144 (0.95) | 346 (0.92) | Ref | |||||
| G | 8 (0.05) | 30 (0.08) | 0.641 | 0.287-1.431 | 1.20 | 0.27430 |
Table 5: Genotype frequencies of MAGEA1 gene polymorphism in colorectal cases and controls (tumour location: colon or rectum)
| SNP | Population | Genotype frequency (N) | Allele frequency | X2 | P value | |||
|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | T | C | ||||
| rs3788745 | CRS (n=190) | 0.921 (175) | 0.058 (11) | 0.021 (4) | 0.950 | 0.050 | - | - |
| HCB (n=90) | 0.400 (36) | 0.244 (22) | 0.356 (32) | 0.522 | 0.478 | 73.41313 | <0.005 | |
| JPT (n=88) | 0.409 (36) | 0.182 (16) | 0.409 (36) | 0.500 | 0.500 | 78.3675 | <0.005 | |
| YRI (n=120) | 0.750 (90) | 0.133 (16) | 0.117 (14) | 0.817 | 0.183 | 14.26769 | <0.005 | |
| rs3788749 | CRS (n=185) | 0.962 (178) | 0.038 (7) | - | 0.981 | 0.019 | - | - |
| CEU (n=120) | 0.100 (12) | 0.100 (12) | 0.800 (96) | 0.150 | 0.850 | 222.2001 | <0.005 | |
| HCB (n=v) | 0.067 (6) | 0.178 (16) | 0.756 (68) | 0.156 | 0.844 | 200.6116 | <0.005 | |
| JPT (n=88) | 0.045 (4) | 0.114 (10) | 0.841 (74) | 0.102 | 0.898 | 218.5015 | <0.005 | |
| YRI (n=120) | - | - | 1.000 (120) | - | 1.000 | 290.7495 | <0.005 | |
| rs3788753 | CRS (n=188) | 0.883 (166) | 0.074 (14) | 0.883 (8) | 0.920 | 0.080 | - | - |
| CEU (n=120) | 0.967 (116) | 0.033 (4) | - | 0.983 | 0.017 | 5.524966 | 0.019 | |
| HCB (n=88) | 0.409 (36) | 0.250 (22) | 0.341 (30) | 0.534 | 0.466 | 55.27718 | <0.005 | |
| JPT (n=88) | 0.409 (36) | 0.182 (16) | 0.409 (36) | 0.500 | 0.500 | 62.97618 | <0.005 | |
| YRI (n=120) | 0.750 (90) | 0.133 (16) | 0.117 (14) | 0.817 | 0.183 | 7.389096 | 0.007 | |
Abbreviations: CRS: Saudi population residing in the Riyadh region of central Saudi Arabia. CEU: Utah residents with Northern and Western European ancestry from the CEPH collection. HCB: Han Chinese in Beijing, China. JPT: Japanese in Tokyo, Japan. YRI: Yoruba in Ibadan, Nigeria.
Table 6: MAGEA1 Allele and genotype frequencies in Riyadh region and other populations
The results revealed no significant relationship regarding patients’ ages, whether patients were above or below 57 years old. That is, ages did not appear to affect the genotype and allele allocations of the three MAGE-A1polymorphisms (Table 4). Third, correlations based on tumor locations (the colon or rectum) between colon cancer risks and individual MAGE-A1SNPs were assessed (Table 5). The SNP rs3788749 had a significant correlation with increased CRC risks among those diagnosed with CRC in the rectum. The CT and CT + TT genotype frequencies were similar in that the frequency was 12% for colon cancer patients and 4% for the control subjects. These genotypes increased the risk of CRC development by more than three times in Saudi patents (OR= 3.416, CI= 1.223–9.538, and p= 0.014). Furthermore, in the Saudi CRC population, T alleles were present more than three-fold higher than in the control population: OR= 3.264, CI= 1.193–8.930 and, p= 0.024 (Table 5).
Comparison between Saudi Arabian individuals and those in other ethnics groups
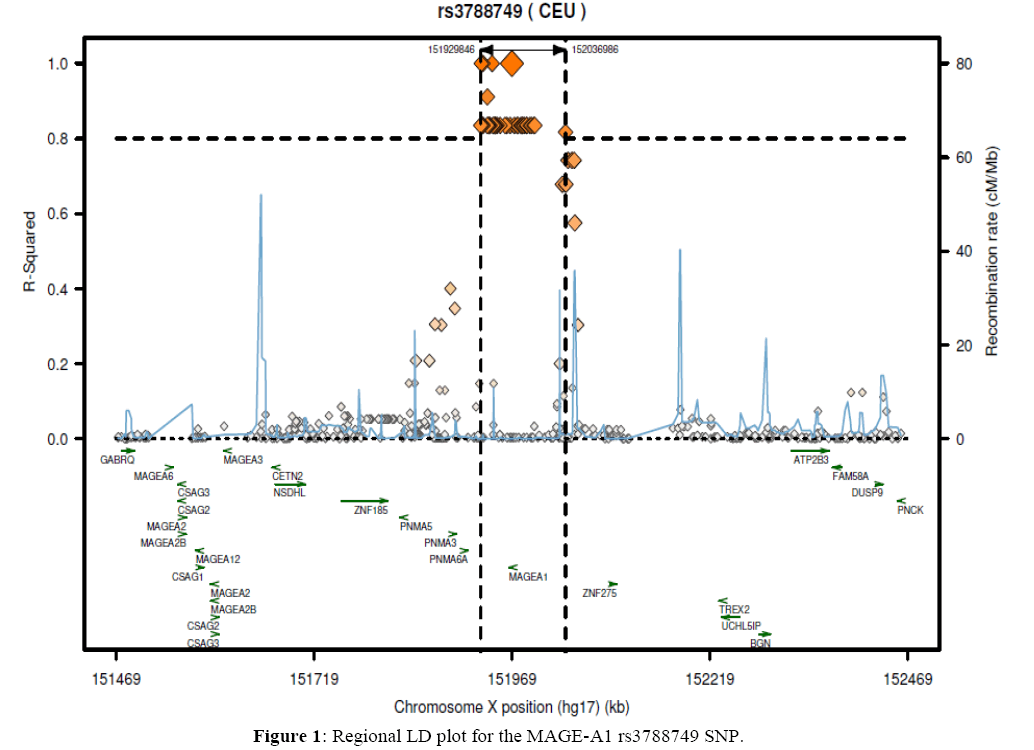
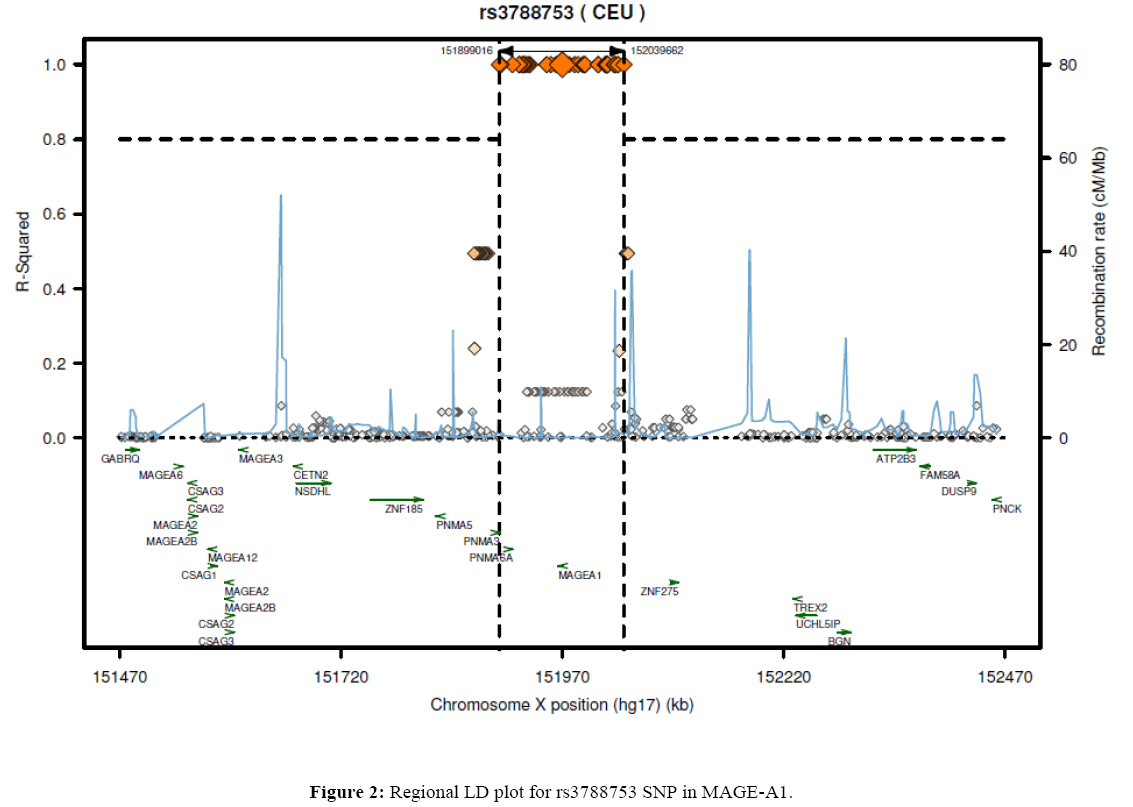
The Saudi Arabian individuals examined in this study were genotyped to compare the samples of the residents of Riyadh region with results in other ethnics groups obtained from the literature (Table 6). We studied 190, 185 and 188 samples from Riyadh region, for the rs3788745, rs3788749, and rs3788753 SNP genotypes, respectively. The allelic and genotypic frequencies for rs3788745 and rs3788749 SNPs were significantly different among Chinese (Han Chinese in Beijing), Japanese (Japanese in Tokyo), Nigerian (Yoruba in Ibadan), Kenyan (Maasai in Kinyawa), and Utah residents of Northern and Western European ancestry for rs3788749 compared to those of the Saudi Arabian population (P<0.005). For MAGE-A1rs3788753, the Saudi population (CRS) was significantly different from Chinese (HCB), Japanese (JPT), Nigerian (YRI), Kenyan (MKK), Utah residents of Northern and Western European ancestry (CEU) populations. Moreover, as presented in Figures 1 and 2, regional linkage disequilibrium (LD) plots were constructed using SNP annotation and Proxy Search (SNAP, http://www.broadinstitute.org/mpg/snap/ldplot.php). The maximum r2 values for the determined SNPs were 0.911 for rs3788749 and 0.494 for rs3788753. Nevertheless, there was no LD data found in the literature for rs3788745.
Abbreviations: LD: Linkage Disequilibrium; MAGE: Melanoma Associated Antigen; SNP: Single-Nucleotide Polymorphism; CEU: Utah Residents with Northern and Western European Ancestry from the CEPH Collection; CEPH: Centre D”Etude Du Polymorphism Human.
Abbreviations: LD: Linkage Disequilibrium; MAGE: Melanoma Associated Antigen; SNP: Single-Nucleotide Polymorphism; CEU: Utah Residents with Northern And Western European Ancestry from the CEPH Collection; CEPH: Centre D”Etude Du Polymorphism Human.
Discussion
Susceptibility to CRC, a multifactorial disease, arises due to both environmental and genetic factors (Park Y et al., 2016). This cancer affects both males and females, and the CRC incidence has increased in Saudi Arabia in recent years. The early detection of CRC can allow patients to receive proper treatment, thereby reducing CRC morbidity, mortality, and incidence (Tan SC, 2018); however, appropriate screening methods are crucial for this detection. The occurrence of CRC is commonly associated with genetic polymorphisms in different genes (Ulrich CM, Robien K, McLeod HL, 2003), such as PITX1 (Gunathilake MN et al., 2018) and TLR-9 (Semlali A et al., 2016). One gene, MAGE-A1, is not expressed in normal tissues except the testis, but it is expressed in tumor tissues or tumor cells (Vanderbruggen P et al., 1991; Feichtinger J et al., 2012).
This unique expression pattern and the antigenic properties of the protein expressed by MAGE-A1allow this gene to be used as an immunotherapeutic target and cancer biomarker (Lee AK and Potts PR, 2017). The main hypotheses of the present study were that MAGE-A1is important in cancer initiation and that its genetic changes are associated with increased CRC risks. Consequently, the principal objective of this study was to investigate the effects of gene polymorphisms, specifically rs3788753, rs3788749, and rs3788745, and their associations with increased CRC risks in Saudi patients.
The results for MAGE-A1genetic polymorphism revealed no relationship between the allelic and genotypic distributions of the MAGE-A1SNPs rs3788745 and rs3788753 regarding CRC risks in the Saudi population. However, a significant relationship was evident between minor genetic and allelic allocations in the MAGE-A1SNP rs3788749 in terms of CRC risks in Saudi patients. According to the NCBI’s SNP database, rs3788749 is an intron SNP, and it may increase MAGE-A1gene expression through its independent functioning as a binding site for transcription factors (Deng N, Zhou H, Fan H, Yuan Y, 2017; Zhu X, Asa SL, Ezzat S, 2009). Nevertheless, many recent studies have reported that this “intron-mediated enhancement” can lead to increases in gene transcription, gene stability, and the efficiency of mRNA translation. For example, Zhu et al (2009) determined that polymorphic sequences in intron 2 of the FGFR 2 gene was related to a constitutive histone acetylation at multiple sequences in this intron that harbored putative transcription binding sites (Zhu X, Asa SL, Ezzat S, 2009).
Significant correlations were found between the rs3788749 SNP and cancer of the rectum, but no significant correlations were found between the SNP and cancer of the colon at the genetic and allelic levels. This finding suggested that this polymorphism is a specific marker for rectal cancer and therefore could be useful as a rectal cancer biomarker for the early detection of CRC in Saudi patients. By contrast, the three MAGE-A1SNP locus genotypes showed no correlation with clinical parameters, such as gender and age, in Saudi males and females.
No previous studies have reported an association between MAGE-A1genetic polymorphisms and CRC susceptibilities. Wang et al. (2004) demonstrated an association between the distribution of MAGE-A1SNPs and hepatocellular carcinoma in a Chinese population, but the tested SNPs were not the same as those examined in the present study (Wang LP et al., 2004). A literature search for the MAGE-A1SNPs rs3788745 and rs3788749 revealed a similar profile for the Riyadh populations and other ethnic groups and a similar profile to that described for SNPs in other genes, such as TLR4 (Kohailan M et al., 2017) and TLR6 (Kohailan M et al., 2016). At the expression level, different studies have shown a higher expression of the MAGE-A1gene in colon cancer tissues than in normal colon tissues (Feichtinger J et al., 2012; Hofmann O et al., 2008) ; Hilal NR, Novikov DV, Novikov VV, Karaulov AV, 2017).
The MAGE-A family members, such as the MAGE-A1gene, could be involved in the progression of gastric cancer, as determination of their expression profiles appears to have prognostic value for patients with gastric cancer (Lian Y et al., 2017). To date, no previous studies have examined the relationship between the genetic variations of the MAGE-A1rs3788753, rs3788749, and rs3788745 SNPs among Saudi patients with CRC. Consequently, the strength this work is its novelty; it is the first study that validates the correlation between genetic polymorphism of the MAGE-A1gene and CRC development risk, particularly in the Saudi ethnic population, who suffer from several genetic diseases due to consanguinity. However, this work also has some limitations. First, the number of samples is small; therefore, larger sample sizes and studies of other ethnic populations are required to confirm the associations between CRC progression and MAGE-A1SNPs, especially SNPs located in regulatory regions of the gene. Second, studies are also needed on the function of these genetic variants on CRC development; therefore, we propose to conduct future investigations of the effect of these MAGE-A1SNPs on CRC cell proliferation, apoptosis, and migration in order to understand the mechanisms of action of these polymorphisms in the regulation of CRC development.
Conclusion
The genotypes and alleles of the rs3788749 MAGE-A1SNP were associated with CRC susceptibility in the Saudi population. This suggests that this polymorphism may serve as a rectal cancer biomarker for the early detection of CRC. However, further investigations are necessary to determine the mechanisms and the functional aspects this relationship between rs3788749 polymorphisms and CRC.
Acknowledgment
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the Research Project No R5-16-03-42.
Disclosure
The authors report no conflicts of interest in this work.
About the Authors
Corresponding Author
Abdelhabib Semlali
Groupe de Recherche en Écologie Buccale, Département de stomatologie, Faculté de Médecine Dentaire, Université Laval, Québec, Qc, Canada
- Email:
- asemlali@ksu.edu.sa
References
- Kruk J and Czerniak U (2013) Physical activity and its relation to cancer risk: updating the evidence. Asian Pac J Cancer Prev 14: 3993-4003. https://doi.org/10.7314/apjcp.2013.14.7.3993
- Peters U, Jiao S, Schumacher FR, Hutter CM, et al. (2013) Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology 144:799.
- Al-Thafar AK, Al-Naim AF, Albges DS, Boqursain SK, et al. (2017) Knowledge Attitude and Practice of Colorectal Cancer among School Teachers in Al-Ahsa Saudi Arabia. Asian Pac J Cancer Prev. 18:2771-4.
- Ibrahim EM, Zeeneldin AA, El-Khodary TR, Al-Gahmi AM, et al. (2008) Past, present and future of colorectal cancer in the Kingdom of Saudi Arabia. Saudi J Gastroenterol 14:178-182. https://doi.org/10.4103/1319-3767.43275
- Krupa R, Sliwinski T, Wisniewska-Jarosinska M, Chojnacki J, et al. (2011) Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer--a case control study. Mol Biol Rep. 38:2849-2854.
- Lurje G, Zhang W, Lenz HJ (2007) Molecular prognostic markers in locally advanced colon cancer. Clinical Colorectal Cancer 6: 683-690. https://doi.org/10.3816/ccc.2007.n.037
- Peltekova VD, Lemire M, Qazi AM, Zaidi SHE, et al. (2014) Identification of genes expressed by immune cells of the colon that are regulated by colorectal cancer- associated variants. International Journal of Cancer 134: 2330-2341. https://doi.org/10.1002/ijc.28557
- Ulrich CM, Robien K, McLeod HL (2003) Cancer pharmacogenetics: Polymorphisms, pathways and beyond. Nature Reviews Cancer 3: 912-920. https://doi.org/10.1038/nrc1233
- Semlali A, Parine NR, Al Amri A, Azzi A, et al. (2016) Association between TLR-9 polymorphisms and colon cancer susceptibility in Saudi Arabian female patients. Oncotargets and Therapy 10:1-11. https://doi.org/10.2147/ott.s106024
- Chen XQ, Mao JY, Li WB, Li J, et al. (2017) Association between CYP24A1 polymorphisms and the risk of colonic polyps and colon cancer in a Chinese population. World J Gastroenterol 23: 5179-5186. https://doi.org/10.3748/wjg.v23.i28.5179
- Alharithy RN (2014) Polymorphisms in RETN gene and susceptibility to colon cancer in Saudi patients. Ann Saudi Med 34: 334-339. https://doi.org/10.5144/0256-4947.2014.334
- Kapitanovic S, Cacev T, Catela Ivkovic T, Loncar B, et al.(2014) TNFalpha gene/protein in tumorigenesis of sporadic colon adenocarcinoma. Exp Mol Pathol 97: 285-291. https://doi.org/10.1016/j.yexmp.2014.08.003
- Deng N, Zhou H, Fan H, Yuan Y (2017) Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 8: 110635-110649. https://doi.org/10.18632/oncotarget.22372
- Stevanovic S (2002) Identification of tumour-associated T-cell epitopes for vaccine development. Nature Reviews Cancer 2: 514-520. https://doi.org/10.1038/nrc841
- Babatunde KA, Najafi A, Salehipour P, Modarressi MH, et al. (2017) Cancer/Testis genes in relation to sperm biology and function. Iran J Basic Med Sci. 20: 967-974. https://doi.org/10.1038/nrc841
- Scanlan MJ, Simpson AJ, Old LJ (2004) The cancer/testis genes: review, standardization, and commentary. Cancer Immun 4:1.
- Caballero OL and Chen YT (2009) Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci 100: 2014-2021. https://doi.org/10.1111/j.1349-7006.2009.01303.x
- Simpson AJG, Caballero OL, Jungbluth A, Chen YT, et al. (2005) Cancer/testis antigens, gametogenesis and cancer. Nature Reviews Cancer 5: 615-625. https://doi.org/10.1038/nrc1669
- Vanderbruggen P, Traversari C, Chomez P, Lurquin C, et al. (1991) A Gene Encoding an Antigen Recognized by Cytolytic Lymphocytes-T on a Human-Melanoma. Science 254: 1643-1647. https://doi.org/10.1126/science.1840703
- Castelli C, Rivoltini L, Andreola G, Carrabba M, et al. (2000) T-cell recognition of melanoma-associated antigens. J Cell Physiol 182: 323-331. https://doi.org/10.1002/(sici)1097-4652(200003)182:3%3C323::aid-jcp2%3E3.0.co;2-#
- Katsura Y and Satta Y (2011) Evolutionary history of the cancer immunity antigen MAGE gene family. PLoS One 6:e20365. https://doi.org/10.1371/journal.pone.0020365
- Doyle JM, Gao J, Wang J, Yang M (2010) MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell 39: 963-974. https://doi.org/10.1016/j.molcel.2010.08.029
- Chomez P, De Backer O, Bertrand M, De Plaen E,et al. (2001) An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res 61: 5544-5551.
- Park Y, Lee J, Oh JH, Shin A, et al. (2016) Dietary patterns and colorectal cancer risk in a Korean population: A case-control study. Medicine 95: e3759. https://doi.org/10.1097/md.0000000000003759
- Tan SC (2018) Low penetrance genetic polymorphisms as potential biomarkers for colorectal cancer predisposition. Journal of Gene Medicine 20: e3010. https://doi.org/10.1002/jgm.3010
- Gunathilake MN, Lee J, Cho YA, Oh JH, et al. (2018) Interaction between physical activity, PITX1 rs647161 genetic polymorphism and colorectal cancer risk in a Korean population: a case-control study. Oncotarget 9:7590-7603. https://doi.org/10.18632/oncotarget.24136
- Feichtinger J, Aldeailej I, Anderson R, Almutairi M, et al. (2012) Meta-analysis of clinical data using human meiotic genes identifies a novel cohort of highly restricted cancer-specific marker genes. Oncotarget 3:843-853. https://doi.org/10.18632/oncotarget.580
- Lee AK and Potts PR (2017) A Comprehensive Guide to the MAGE Family of Ubiquitin Ligases. Journal of Molecular Biology 429: 1114-1142. https://doi.org/10.1016/j.jmb.2017.03.005
- Zhu X, Asa SL, Ezzat S (2009) Histone-acetylated control of fibroblast growth factor receptor 2 intron 2 polymorphisms and isoform splicing in breast cancer. Mol Endocrinol 23: 1397-1405. https://doi.org/10.1210/me.2009-0071
- Wang LP, Chen HS, Mei MH, Qin LL, et al. (2004) The genetic polymorphism of melanoma-associated antigen 1 in Chinese normal donors and hepatoma patients. European Journal of Clinical Pharmacology 12: 151-155.
- Kohailan M, Alanazi M, Rouabhia M, Al Amri A, et al. (2017) Two SNPs in the promoter region of Toll-like receptor 4 gene are not associated with smoking in Saudi Arabia. Oncotargets and Therapy10: 745-752. https://doi.org/10.2147/ott.s111971
- Kohailan M, Alanazi M, Rouabhia M, Alamri A, et al. (2016) Effect of smoking on the genetic makeup of toll-like receptors 2 and 6. Oncotargets and Therapy 9: 7187-7198. https://doi.org/10.2147/ott.s109650
- Hofmann O, Caballero OL, Stevenson BJ, Chen YT, et al. (2008) Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci 105: 20422-20427. https://doi.org/10.1073/pnas.0810777105
- Hilal NR, Novikov DV, Novikov VV, Karaulov AV (2017) Cancer-testis genes in colon cancer. Ter Arkh 89: 113-117. https://doi.org/10.17116/terarkh2017895113-117
- Lian Y, Sang M, Gu L, Liu F, et al. (2017)MAGE-A family is involved in gastric cancer progression and indicates poor prognosis of gastric cancer patients. Pathol Res Pract 213: 943-948. https://doi.org/10.1016/j.prp.2017.05.007
Keywords:
Download:
Full PDF- Share This