Molecular Characterization and Differential Interaction of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans with Common Beans (Phaseolus vulgaris)
Received: December 14, 2017
Accepted: January 08, 2018
Published: January 25, 2018
Genet.Mol.Res. 17(1): gmr16039871
DOI: 10.4238/gmr16039871
Abstract
The common bacterial blight, caused by Xanthomonas axonopodis pv. phaseoli (Xap) and X. fuscans subsp. fuscans (Xff), is one of the most important dry bean diseases of bacterial etiology. The present work aimed to study the genetic structure among and within populations, and also identify the pathogenic variability of Xap and Xff isolates evaluating the horizontal and vertical resistance of the host. A total of 42 Xap and Xff isolates were obtained were submitted to molecular characterization by rep-PCR markers, and inoculated in the genotypes CNFP 10120, BRS Notável, and Rosinha G-2, for differential interaction studies. In the analysis of genetic structure, the coefficient of genetic differentiation (GST=0.5154) revealed diversity both within and among the pathogenic populations, confirming the result obtained by AMOVA (FST=0.5198). Of a total of 51 loci, 98% were polymorphic, among Xap 59% and within Xff only 21% were polymorphic. In studies of pathogenic variability, diallelic analyses showed significant effects of the general combination ability (GCA), of the general aggression ability (GAA), and of the specific interaction ability (SIA). Therefore, seven races were classified through the differential behavior of all isolates in the three genotypes of bean.
Introduction
The causal agents of common bacterial blight (CBB), Xanthomonas axonopodis pv. phaseoli (Xap) (Smith) Dye (Young et al. 1996) and X. fuscans subsp. fuscans (Schaad et al. 2005) (Xff) are differentiated phenotypically by in vitro melanin production by Xff. Among the diseases of bacterial origin, CBB deserves special attention due to its wide distribution, ability to reduce production significantly, low efficiency of chemical control, and difficulty of obtaining resistant commercial cultivars (Manten, 2008).
Strategies of control through genetic resistance are researched but obtaining varieties resistant to the disease is hampered primarily due to the high variability regarding pathogenicity and virulence (Kimati et al. 2005). Efficient utilization of genetic resistance in developing cultivars with durable and broad-spectrum resistance requires knowing the sources of resistance in the host species and the genetic diversity of the pathogen populations.
In studies of genetic diversity, a technique that is widely used is the analysis of repetitive regions by means of amplification by rep-PCR (Louws et al. 1995a; Louws et al. 1995b), through the use of three families of repetitive sequences, REP, ERIC, and the BOX element (Martin et al. 1992). According to Alves et al. (2003), these markers are complementary to the repetitive DNA sequences highly conserved and present in multiple copies in the bacterial genome. This technique was used successfully in diversity studies, as well as for identification and classification of several bacteria including Xap (De Bruijn, 1992; Restrepo et al. 2000; López et al. 2006; Mahuku et al. 2006; Munhoz et al. 2011).
Genetic improvement for CBB resistance is genetically complex and of difficult implementation, due to genetic resistance being a character of oligogenic or polygenic inheritance, as reported by some authors (Coyne & Schuster, 1974; Santos et al., 2003). Partial diallel analysis proposed by Melo & Santos (1999), adapted from Griffing’s model IV (Griffing, 1956), and is a methodology able to provide information on the horizontal and vertical resistance of the host and of the presence of virulence and/or aggressiveness of the pathogen. Based on this information, the present study had the following objectives: establish a collection of Xap and Xff isolates from dry bean producing regions in Brazil; study the diversity and genetic structure among and within the populations sampled; evaluate the efficiency of molecular markers in distinguishing the two pathogens; relate the genetic diversity of the pathogen with its geographical distribution; identify the pathogenic variability and differential interaction of Xap and Xff isolates evaluating the horizontal and vertical resistance of the host.
Materials and Methods
Collection, isolation, and pathogenicity test
Collections were carried out in the regions of Goiás, São Paulo, Paraná and Rio Grande do Sul from 2007 to 2011. The plant material with visible symptoms of CBB was transported to the Plant Pathology Laboratory of Embrapa Arroz e Feijão, for the isolation of the bacterium and subsequent detection of the pathogen. The isolation of the bacterium was carried out with the infected bean leaves, following the protocol proposed by Romeiro (2011). Yellow colonies, typical of Xap, obtained from the isolation, were selected and grown from a single colony in YDC culture medium to obtain pure colonies. All isolates were tested for pathogenicity in two cultivars susceptible to bacterial blight, Rosinha G-2 and Waff 75. Ten-day-old plants, grown in pots in greenhouse conditions, were submitted to inoculation using the method of incision of leaves, described by Rava (1984). The severity was observed for ten days after inoculation through a diagrammatic scale with grades from 0 to 6 (Rava (1988)). In which, note 0 for the absence of symptoms; note 1 for small yellowing in the cut; note 2 for yellowing extending to the region between cuts; note 3 for slight necrosis in the cut region; note 4 for necrosis in the cut region and yellowing in every region between cuts; note 5 for the same aspect noted in the previous note, but with yellowing extending in the region above the upper cut and below the lower cut; and note 6 for completely damaged leaf.
Colony morphology
For cultural characterization, each isolate was grown in triplicate in YDC culture medium at 25°C and evaluated 72 hours after subculturing, observing the characteristics of the colony: smooth and mucoid growth, convex and bright colonies, and brown pigmentation production by colonies of Xff, according to the methodology of Birch et al. (1997).
Extraction bacterial DNA
DNA extraction and purification were carried out with the aid of DNA purification Kit Wizard® (Promega Corp., Madison, WI), following the manufacturer’s Protocol. After extraction, the DNA was quantified in spectrophotometer (NanoDrop ND-1000 UV-Vis Espectofotômetro, NanoDrop Technologies), also analyzing its quality.
Specific detection of Xap and Xff
PCR for detection of Xap and Xff was carried out according to the procedures described by Audy et al. (1994) and Birch et al. (1997) with modifications. The reaction was performed with the multiplex PCR Kit (QIAGEN®) in final volume of 10 μL containing: 5 μL of mastermix solution; 1 μL of Q-solution; 0.3 μM of each primer Xf 1.2 (5’ -ACG CAA GAC CCA TCG TCA TTC -3’) and Xf 2.2 (5’ - ATG GCT CAA GGA AAA ACT TTC AGG -3’); 100 μM of each primer X4c (GGC AAC ACC CGA TCC CTA AAC AGG) and X4e (CGC CGG AAG CAC GAT CCT CGA AG); and 2 μL of DNA (25 ng/μL). The reactions were submitted to 37 cycles of 94°C/30 sec, 65°C/30 sec, 72°C/1 min, and a final extension at 72°C/10 min, in thermal cycler (MJ research, model PTC-100). The positive control was represented by a pathogenic isolate (BRM 25302), while sterile water and isolates that are pathogenic to other hosts, X. axonopodis pv. manihotis (Xam), X. citri subsp. citri (Xcc), X. gardneri (Xg) and X. axonopodis pv. glycines (Xag) were used as negative control. Detection was performed by electrophoresis in 1.5% agarose gel, with Tris-acetate base - EDTA (TAE)0.75x, for 2 hours at 100 Volts. The marker used to estimate the size of the amplified fragments was of 1 kb (Fermentas) and the gel was submitted to bath in a 0.5 mg/l ethidium bromide solution for 15 min, for visualization of bands and photodocumenting.
Genetic characterization of isolates by rep-PCR
The markers ERIC (eric1R 5’ ATG TAA GCT CCT GGG GAT TCA C 3’ and eric 2 5 ‘AAG TAA GTG ACT GGG GTG AGC G 3’), BOX (boxA1R 5’ CTA CGC CAA GGC GAC GCC TGA CG 3’) and REP (rep1 R-I 5’ III CGI CGI CAT CIG GC 3’ and rep2 R-I 5’ ICG ICT TAT GIG GCC TAC 3’) were used separately for amplification of the DNA of the isolates. The reaction was carried out with the aid of multiplex PCR Kit (QIAGEN®) in a final volume of 10 μL containing 5 μL of mastermix solution, 1 μL of Q-solution, 50 pmol for each initiator, supplementing to the final volume with sterile and RNAse-free water. The reactions were submitted to the following cycles: for the initiator ERIC, an initial cycle at 95°C for 7 min; 30 cycles of denaturation at 94°C for 1 min, girdling at 44°C for 1 min, and extension at 65°C for 8 min followed by a final extension cycle at 65°C for 15 min. For the initiators BOX and REP, the steps of amplification: an initial cycle at 95°C for 7 min; 35 cycles of denaturation at 94°C for 1 min, girdling at 55°C for 1 min (40°C for 1 min for REP), and extension at 65°C for 8 min, followed by a final extension cycle at 65°C for 15 min. After amplification, we added 5 μl of bromophenol blue dye and 5 μL of SYBR gold, to the amplified product, and, then, we conducted electrophoresis in 1.5% agarose gel, prepared with Tris-acetate base - EDTA (TAE) 0.75x, for 7 hours at 50 Volts.
Data analysis
For the results obtained by rep-PCR, we conducted a haplotype analysis, in which each locus by initiator was identified according to estimated molecular weight receiving a designation (present loci received value 1 and absent loci received 0 value), considering fragments of identical molecular weight as homologs, and eliminating low-resolution fragments. Thus, each isolate received a combined binary haplotype, in which all amplified loci were represented. To ensure data reliability, two isolates (replicas) were amplified and loaded at the end of each gel. The frequency of haplotypes was calculated for each region of collection. To build the dendrogram, we used the similarity matrix generated in the UPGMA algorithm (method of non-weighted arithmetic means) in the DARwin 5.0 computer program. The coefficient of similarity used in such procedure was that of Jaccard. To determine the consistency of the nodes of the dendrogram generated, 10,000 bootstrappings were performed.
Genetic structure
The isolates were divided into three populations, with the first consisting of Xap isolates, the second of Xff isolates, and the third of isolates pathogenic to other hosts (Xam, Xg, Xag e Xcc). Based on the binary data obtained by rep-PCR, the genotypic diversity of the populations was estimated by the Shannon-Wiener indices.
Through the POPGENE 1.31 program (Yeh et al. 1999), we obtained the percentage of polymorphic loci (P), coefficient of genetic diversity of Nei (Gst) (Nei, 1973), the total genetic diversity (Ht), and the genetic identity of each population.
AMOVA, used to estimate the variance components, was conducted in the Arlequin program (Excoffier et al. 2005), in which the total variance among the isolates was subdivided into variance among and within the two populations (Xap and Xff).
Differential interaction between dry bean genotypes and Xap and Xff isolates
The test was conducted in a greenhouse in entirely random design with four replicates and negative control. There were 21 Xap isolates and 21 Xff isolates, inoculated in three genotypes, CNFP 10120, BRS Notável, and Rosinha G-2. The bacterial suspension was adjusted to a concentration of 108 CFU/mL and inoculated in ten-day-old plants, through the method of incision of leaves, described by Rava (1984). The severity was evaluated ten days after inoculation through a diagrammatic scale with grades from 0 to 6 (Rava (1988)).
The mean severity grades were submitted to analysis of variance and subsequently submitted to analysis of general combination ability (GCA), general aggression ability (GAA), and specific interaction ability (SIA), calculated according to the method proposed by Melo and Santos (1999). For that, Group I was composed of three genotypes, and Group II was composed of 42 pathogenic isolates. In pathogen-host interactions, GCA represented horizontal resistance of the genotypes to the different isolates of the pathogen or physiological races. The GAA of the pathogen represented the aggressiveness of each physiological race when inoculated with the genotypes of the host. The SIA indicated the interaction among the components of the two groups. This estimate represents virulence of the physiological race and vertical resistance of the genotype.
Results
Collection, isolation and pathogenicity test
All isolates were purified and deposited in the Embrapa’s collection of multifunctional microorganisms. When inoculated in susceptible cultivars, were pathogenic, showing visual symptoms with grades ranging from 3 to 6 (degree of symptoms previously described).
Detection with specific primers
In the multiplex reaction (X4c/X4e and Xf1/Xf2), we obtained only a fragment of 730 pb for the Xap isolates, while the Xff isolates presented two fragments, the first of 730 pb (amplified by the primers X4c/X4) and the second of 450 pb (amplified by the primers Xf1/Xf2). In addition, no amplification was observed in the negative control (water), as well as for the isolates of Xam, Xcc, Xg, and Xag, demonstrating the specificity of these markers.
Colony morphology
All isolates presented smooth and mucoid growth, convex and bright colonies. The production of pigments, observed five days after sub-culturing in YDC, King B, and MNBY culture media, enabled identifying the Xap and Xff isolates. A strong brown coloration was observed in the King B culture medium, after two to three days of culture, while a less intense coloration was observed in the YDC and MNBY media after five to seven days of growth. The isolates belonging to a species of brown pigment (Xff), BRM 25315, BRM 25334, BRM 25349, BRM 25342, BRM 25354 e BRM 27257, did not produce the pigmentation expected for the YDC medium, even after 15 days of growth, still being yellow.
Genetic characterization by rep-PCR
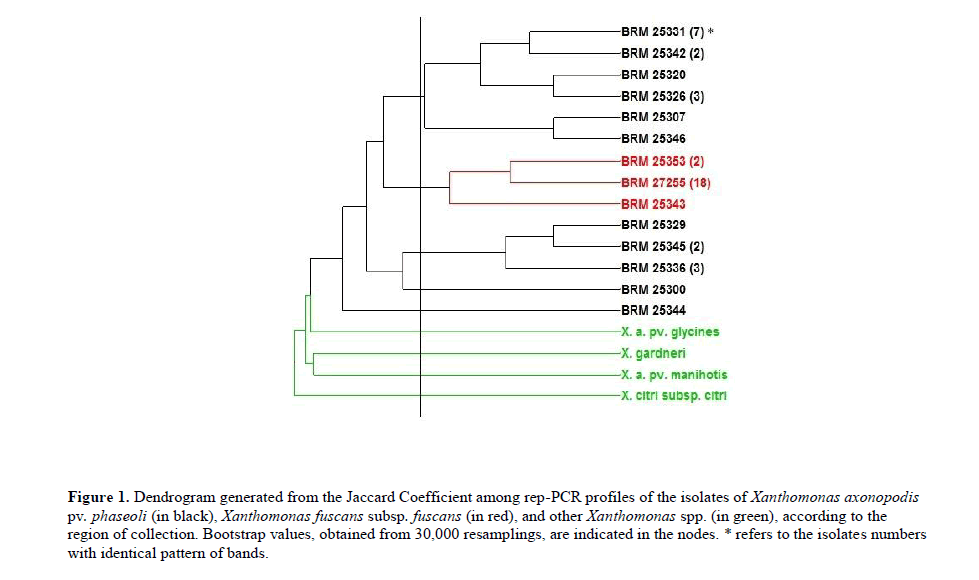
The dendrogram generated (Figure 1) revealed three distinct groups among the isolates belonging to the species causing CBB in common bean. The first group consisted only of isolates of Xap, the second with all isolates of Xff, revealing the presence of clonal isolates in this species for this molecular marker. Since, of the total of 21 isolates Xff, 18 presented identical pattern of bands for these markers. The third group consisted of Xap isolates that showed a clear genetic distance from the remaining isolates of the same species.
Figure 1: Dendrogram generated from the Jaccard Coefficient among rep-PCR profiles of the isolates of Xanthomonas axonopodis pv. phaseoli (in black), Xanthomonas fuscans subsp. fuscans (in red), and other Xanthomonas spp. (in green), according to the region of collection. Bootstrap values, obtained from 30,000 resamplings, are indicated in the nodes. * refers to the isolates numbers with identical pattern of bands.
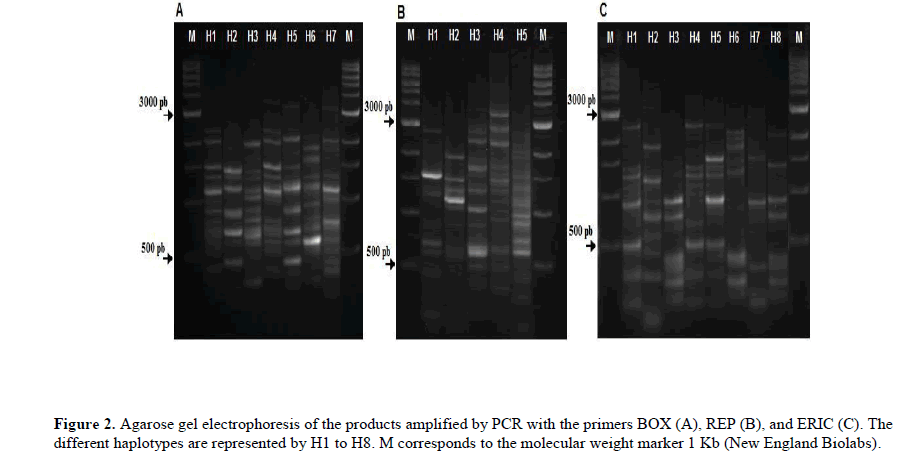
The isolates BRM 25300 and BRM 25344 showed to be more genetically related with non-patovar phaseoli isolates (X. axonopodis pv. manihots, X. gardineli, X. axonopodis pv. glycines, and X. citri subsp. citri). In order to assess the diversity, we also compared haplotypes generated by the rep-PCR amplification. The electrophoretic pattern of Xap and Xff generated seven, five, and eight haplotypes for markers BOX, REP, and ERIC, respectively (Figure 2).
These haplotypes, when analyzed together, formed a total of 12 haplotypes combined between the Xap and Xff isolates (Table 1).
| Origin | Isolate | BOX | REP | ERIC | Haplotype Combined |
|---|---|---|---|---|---|
| BRM 25297 (Xap) | H1 | H1 | H1 | HC1 | |
| BRM 25302 (Xap) | H1 | H1 | H1 | HC1 | |
| BRM 25300 (Xap) | H4 | H2 | H2 | HC2 | |
| BRM 25301(Xap) | H1 | H1 | H1 | HC1 | |
| BRM 25305 (Xap) | H1 | H1 | H1 | HC1 | |
| BRM 25306 (Xap) | H1 | H1 | H1 | HC1 | |
| BRM 25307 (Xap) | H1 | H1 | H4 | HC4 | |
| BRM 25314 (Xap) | H1 | H1 | H1 | HC1 | |
| BRM 25321 (Xap) | H7 | H1 | H1 | HC6 | |
| BRM 25304 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25308(Xff) | H2 | H3 | H3 | HC3 | |
| Paraná | BRM 25347(Xff) | H2 | H3 | H3 | HC3 |
| BRM 25348 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25350 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25351(Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25352 (Xff) | H2 | H3 | H8 | HC12 | |
| BRM 25353(Xff) | H2 | H3 | H8 | HC12 | |
| BRM 25354 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25355 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25356 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25357(Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25359 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25326 (Xap) | H7 | H1 | H1 | HC6 | |
| BRM 25328 (Xap) | H3 | H1 | H1 | HC7 | |
| BRM 25329 (Xap) | H6 | H1 | H1 | HC8 | |
| BRM 25331(Xap) | H1 | H1 | H1 | HC1 | |
| Goiás | BRM 25315 (Xff) | H2 | H3 | H3 | HC3 |
| BRM 25316 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25349 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 27255 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 27257 (Xff) | H2 | H3 | H3 | HC3 | |
| BRM 25319 (Xap) | H1 | H1 | H1 | HC1 | |
| BRM 25320 (Xap) | H3 | H1 | H5 | HC5 | |
| BRM 25342 (Xap) | H1 | H1 | H1 | HC1 | |
| Rio Grande do Sul | BRM 25343 (Xff) | H2 | H5 | H6 | HC10 |
| BRM 25344 (Xap) | H5 | H4 | H7 | HC11 | |
| BRM 25345 (Xap) | H4 | H1 | H1 | HC9 | |
| BRM 25346(Xap) | H1 | H1 | H4 | HC4 | |
| BRM 25333 (Xap) | H4 | H1 | H1 | HC9 | |
| BRM 25336 (Xap) | H4 | H1 | H1 | HC9 | |
| São Paulo | BRM 25334 (Xff ) | H2 | H3 | H3 | HC3 |
| BRM 25337 (Xff) | H2 | H3 | H3 | HC3 |
Table 1. Individual haplotypes BOX, REP, and ERIC, and combined haplotypes of the isolates of Xanthomonas axonopodis pv. phaseoli (Xap) and Xanthomonas fuscans subsp. fuscans (Xff) of the states of Paraná (PR), Rio Grande do Sul (RS), São Paulo (SP), and Goiás (GO).
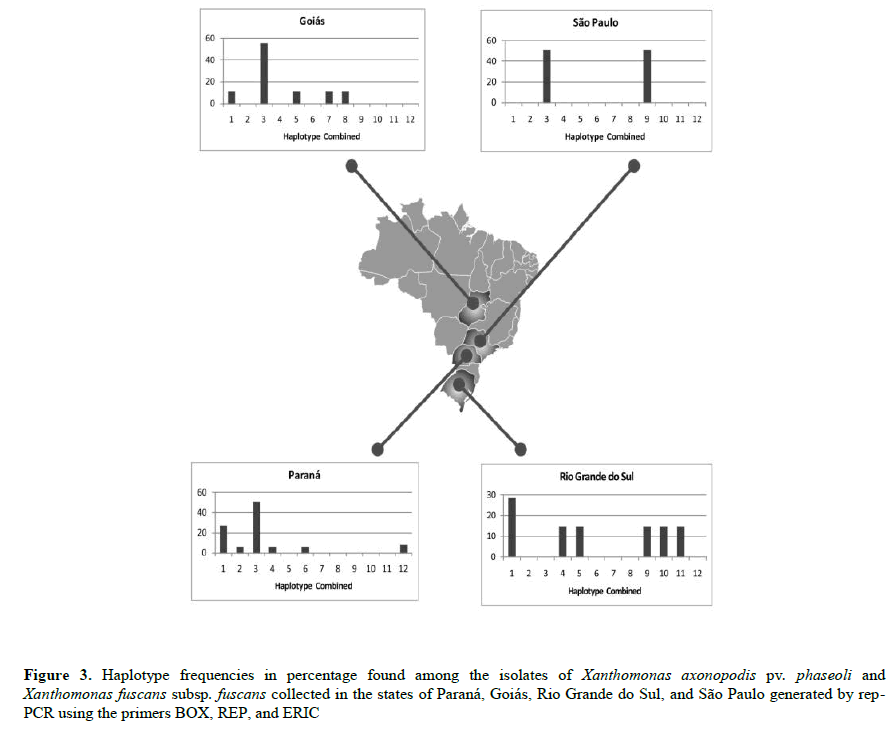
The HC 3, specific to Xff, was the haplotype most commonly identified (42.8% of total isolates), with high frequency in all states, except in Rio Grande do Sul, where it was absent. In Rio Grande do Sul, we also observed the greatest genetic diversity, as each isolate represented a distinct haplotype. The HC 1, specific among the Xap isolates, was the second most frequent (21.4% of total isolates), present in all states, except in São Paulo (Figure 3).
Genetic structure
Of a total of 51 loci, 98% were polymorphic. Among the Xap isolates, 59% of the loci were polymorphic, while among the Xff isolates this measure was of only 21% (Table 2).
| Population | N° of isolates | Polymorphism (%) | Total genetic diversity (Ht) | Shannon index (I) |
|---|---|---|---|---|
| X. a. pv. phaseoli | 21 | 59 | 0, 1376 | 0, 2233 |
| X. f. subsp. fuscans | 21 | 21 | 0, 0352 | 0, 0633 |
| X. sp (Pathogenic to other hosts) | 4 | 47 | 0, 1789 | 0, 2672 |
| All isolates | 46 | 98 | 0, 2385 | 0, 3252 |
Table 2. Percentage of generated polymorphism, total genetic diversity (Ht), and Shannon index for the populations of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans and non-pathogenic to dry bean.
Higher values of the Shannon index (0.22) for Xap revealed that this population was more genetically diverse, confirming the data obtained in the dendrogram. The Nei genetic diversity coefficient revealed genetic differentiation both within and between the two pathogenic populations (GST=0.51). The genetic identity and the Nei genetic distance among the populations revealed that Xap presented higher genetic identity with the non-pathogenic populations (0.86) when compared with Xff (0.79) (Table 3).
| Population | X. a. pv. phaseoli | X. f. subsp. fuscans | Non-pathogenic |
|---|---|---|---|
| X. a. pv. phaseoli | *** | 0,7957 | 0,8611 |
| X. f. subsp. fuscans | 0,2285 | *** | 0,7154 |
| X. sp (Pathogenic to other hosts) | 0,1496 | 0,3349 | *** |
Table 3. Nei measure (1978) for genetic identity (above the diagonal) and genetic distance (below the diagonal) for the three populations of Xanthomonas axonopodis pv. phaseoli, Xanthomonas fuscans subsp. fuscans, and other Xanthomonas spp. (non-pathogenic).
The AMOVA showed that most of the genetic diversity was distributed among the pathogenic populations (51.98%, Table 4).
| Source of variation | d.f. | SS | CV | % Variation |
|---|---|---|---|---|
| Among populations | 1 | 235.851 | 3,80 | 51,98 |
| Within populations | 41 | 146.818 | 3,51 | 48,02 |
| Total | 42 | 382.619 | 7,31 |
Table 4. Analysis of molecular variance (AMOVA) among and within Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans.
Differential interaction between dry bean genotypes and Xap and Xff isolates
Diallelic analyses showed significant effects of general combination ability (GCA) and general aggression ability (GAA), indicating variability for horizontal resistance in the host and for aggressiveness in the pathogen. And there were also significant effects for the specific interaction ability (SIA) revealing the existence of interaction between isolate and genotype of the host.
However, when analysing the developments for the sources of variation (Table 5), it is observed that most of the variation of the mean square was due to the GCA, which indicates the predominance of horizontal resistance. The results indicate that the material with higher horizontal resistance was BRS Notável (GCA=-0.86) and the material with lower horizontal resistance was Rosinha G-2 (GCA=1.38). The most aggressive isolates were BRM 27257, from the state of Goiás, BRM 25300 from the state of Paraná, and BRM 25328 from the state of Goiás.
| Source of variation | d.f. | SS | MS | F |
|---|---|---|---|---|
| Genotypes (G-I) | 2 | 444 | 222 | 1564,151 * |
| Isolates (G-II) | 40 | 249 | 6 | 43, 959 * |
| Interaction (GI x GII) | 80 | 62 | 0,780222 | 5,503 * |
| Residual | 253 | 36 | 0,141772 | |
| General mean: | 36 | |||
| CV (%) | 10, 50 |
*Significant at 5% probability (F test)
Table 5. Analysis of variance of the partial diallel scheme for the CBB severity data in the genotypes inoculated with 42 isolates of X. a.pv. phaseoli and X. f. subsp. fuscans.
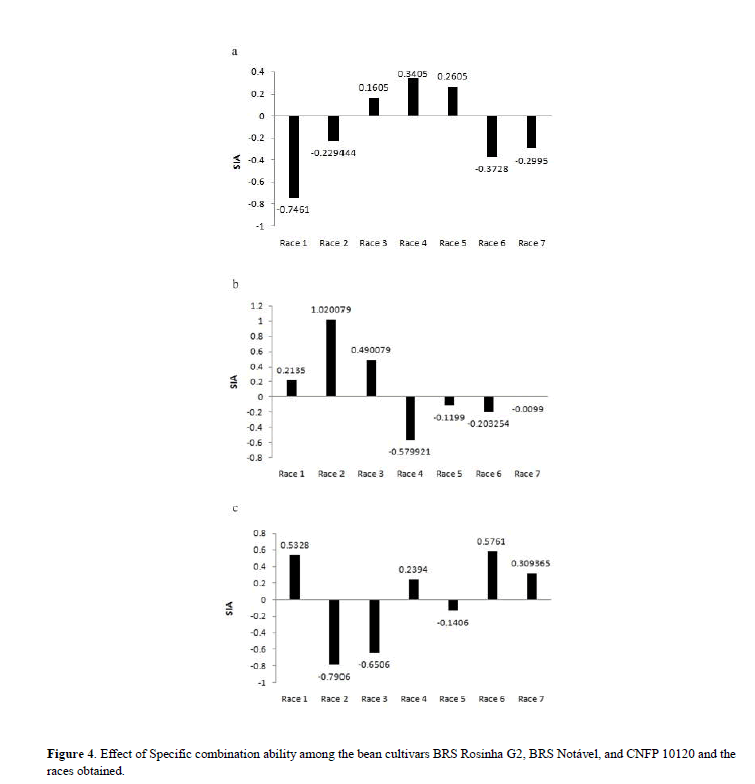
The significance of SIA allowed the stratification of isolates in seven physiological races that presented differential behavior in the three genotypes of dry bean (Figure 4).
Discussion
The pathogens Xanthomonas axonopodis pv. phaseoli and X. fuscans subsp. fuscans presented morphological and genetic differences that allow to differentiate them. Phenotypically, Xff is differentiable due to producing brown pigment in YDC culture medium (Burkholder, 1930; Mkandawire et al. 2004; Schaad et al. 2005). The results regarding phenotypic characterization were better observed when the isolates were grown in King B culture medium, as observed by Birch et al. (1997). As reported by some authors, the time of inoculation on the slides until the production of such pigments can vary substantially (Goodwin & Sopher, 1994; Birch et al. 1997). It is suggested that these variations may occur due to differences in the time taken to produce pigment in each culture medium; thus, the King B medium performed more efficiently for production of brown pigmentation in Xff colonies.
Considering the difficulty of classifying isolates based solely on the color of colonies in different culture media, molecular analyses were performed to complement the phenotypic analyses. The specific markers X4c/X4e, all Xap and Xff isolates presented fragments of same size as the positive control (730 pb). On the other hand, Audy et al. (1994) observed the amplification of fragments of 730 pb for the Xap and Xff isolates, and occasionally an additional fragment of 550 pb only for the Xff isolates, when submitted to girdling temperature of 65°C. Halfeld-Vieira et al. (2001), also did not identify for any Xff isolate an additional fragment of 550 pb, and suggested reducing girdling temperature to 55°C. When combining the set of primers in a multiplex reaction (X4c/X4e and Xf1/ Xf2), it was possible to distinguish the two species of bacteria through the size of the fragment obtained. Boureau et al. (2013) evaluated the effectiveness of the set of primers Xf1/Xf2 in identifying several pathovars of Xanthomonas and observed amplification of a fragment of 450 pb specific only to Xff isolates. It is important to stress the use of the detection primers in conjunction, since only X4c/X4e are not effective to distinguish the two species of Xanthomonas.
The identification, through the rep-PCR profiles, of Xap isolates genetically like other species of Xanthomonas, can be explained through the four different genetic strains that compose the phaseoli pathovar. As reported by Hajri et al. (2009) genotypes belonging to the genetic lineages 2 and 3 of X. axonopodis pv. phaseoli are phylogenetically closer to genotypes that are pathogenic to citrus or cotton, compared with strains belonging to the genetic lineage 1. Therefore, it is possible that the Xap isolates closest to the other Xanthomonas species (Xam, Xg, Xag e Xcc) belong to different lineages within the phaseoli pathovar. However, further molecular phylogeny studies of these isolates are necessary to confirm these results. Analysis of the results generated by rep-PCR showed that there is genetic diversity among the Xap and Xff isolates collected in the states of Goiás, São Paulo, Paraná, and Rio Grande do Sul. The group observed showed that the collection site was not a significant factor in determining the genetic similarity of the isolates, since no clear relation was established between collection site and group composition. This absence of geographical differentiation can be the result of exchange of contaminated seed among producing regions in Brazil, since one of the main means of dissemination of CBB is through seeds.
When analyzing the distribution of genetic diversity among and within populations (Xap and Xff), we observed higher values for polymorphic loci (59%) and Shannon index (0.22) for Xap, revealing that this population was more genetically diverse. The results of this study agree with those obtained by Mkandawire et al. (2004), who analyzed Xap and Xff isolates collected in different regions of the world, such as East Africa, Brazil, Colombia, Guatemala, Puerto Rico, Spain, and United States. And, through analyses with the markers rep-PCR and RFLP, they obtained polymorphism only for Xap, while the electrophoretic pattern of Xff was homogeneous. Differently, López et al. (2006), when analysing the same pathogens, but collected in the northern Spain, found polymorphism among the isolates of Xff with the markers REP and BOX, and only a few polymorphic loci were observed in the profiles generated with ERIC. Moreover, Mahuku et al. (2006) demonstrated that the marker rep-PCR detected 72% of polymorphic bands in Xff populations of South America, Central America, North America, Africa, Europe, and Australia.
The AMOVA demonstrated that the genetic variation within populations of Xap and Xff was 48.02%, and it is possible that this variation is concentrated within the population of Xap, since higher values for Ht, polymorphic loci, and Shannon index were found in this population. The FST value (0.51) observed in AMOVA demonstrates high genetic divergence among the populations analyzed, as, according to Hartl & Clark (2010), FST values higher than 0.25 indicate high genetic differentiation. Genetic structure similar to the present study was also found by Mahuku et al. (2006) for the same pathogens (Xap and Xff), observing high values for GST (56%), index of genetic identity (0.648) and genetic distance (0.434), confirming the distinct nature of these two species of pathogenic bacteria.
Through the analysis of genetic structure of this study, it was possible to observe that Xap and Xff are genetically distinct, in spite of causing the same symptoms on the bean plant. Therefore, further studies on this virulence gene homology in plasmids are needed in order to elucidate how these genetically distinct bacteria produce the same symptoms in bean plants.
In addition to knowing the genetic diversity of causal agents, it is of great importance to understand the pathogenic variability in order to assist the genetic improvement programs. With such objective, the methodology of Melo and Santos (1999) has been used to identify interactions between genotypes and isolates in several pathosystem (Davide & Souza, 2009; Buiate et al. 2010; Pereira et al. 2015). The method is highly informative in determining the horizontal and vertical resistance of strains, as well as the aggressiveness of pathogenic isolates.
In this study, diallelic analyses showed significant effects of the general combination ability (GCA), of the general aggression ability (GAA), and of the specific interaction ability (SIA). Significant values for SIA reveal the existence of interaction between isolate and genotype of the host, which can characterize the existence of vertical resistance, as already reported by some authors for CBB (Coyne & Schuster, 1974; Santos et al. 2003; Manzanera et al. 2006). However, the developments for the sources of variation showed that most of the variation of the mean square was due to GCA, which indicates the predominance of horizontal resistance. Usually both types of resistance occur in conjunction, as observed, for example, by Parlevliet (1981) in assessing the percentage of rice leaf area infected by X. oryzae.
Silva et al. (2000) evaluating the general combination ability through the methodology of unbalanced diallels among 5 genotypes of dry bean and one isolate of Xap, observed that the genotype Rosinha G-2 presented more unfavorable GCA, showing lower capacity to contribute with resistance alleles for the gene set under study, which corroborates the result of this study, in which the genotype Rosinha G-2 proved the material with lowest horizontal resistance (GCA=1.38).
In relation to the isolates, the estimated general aggression ability (GAA) was significant, indicating the occurrence of different aggressiveness alleles. The most aggressive isolates (BRM 27257, BRM 25300, BRM 25328) present higher number of virulence allels and higher value for GAA. Isolates with higher aggressiveness should be preferred for assessment of the level of resistance of genotypes of bean plant. Considering this, the partial diallels methodology of Melo & Santos (1999) enabled the classification of seven races through the differential behavior of 42 isolates of Xap and Xff in three bean plant genotypes. As observed in Figure 4, race 2 presented greater variation in SIA values, proving the most virulent race to the BRS Notável genotype and the less virulent to CNFP 10120. The host BRS Rosinha G-2 presented higher vertical resistance to race 1, while BRS Notável and CNFP 10120, to races 4 and 2, respectively.
The occurrence of interaction between isolates and genotypes indicates the association between virulence and vertical resistance in this pathosystem. Through the study of differential interaction between 6 isolates of X. axonopodis pv. phaseoli and 33 genotypes of bean plant, Silva et al. (2008) observed that the isolate Xap 2 was more pathogenic to the cultivar BRS Esplendor and Xap 4 was less pathogenic, verifying the virulence of the pathogen. In Phaseolus acutifolius, Opio et al. (1996) found differential interaction between the 30 isolates of Xap and 8 genotypes and 8 distinct physiological races of Xap were defined, suggesting the occurrence of gene to gene interaction. Despite this apparent gene to gene interaction, it is necessary to establish a differentiating series of Phaseolus vulgaris genotypes for classification of races of Xap.
Although some authors report the occurrence of pathogenic variability between the two species, in which Xff tends to be more virulent than Xap (Opio et al. 1996; Mutlu et al. 2008; Duncan et al. 2011). In this study, it was clear that there was no difference in the degree of aggressiveness between the species Xap and Xff, showing that despite the two species being genetically distinct, they present the same levels of aggressiveness in the host. Lastly, this is the first work that uses the molecular markers rep-PCR to study the population genetic structure of Brazilian isolates of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans from different bean producing regions. Our studies reveal that the molecular patterns obtained with markers BOX, REP, and ERIC showed that Xap and Xff isolates are genetically distinct; however, we observed no grouping of isolates according to region of collection. About the populational structure, AMOVA showed that most of the genetic variability is among populations, and much of this divergence is concentrated in the population of Xap, since higher values for total gene diversity (Ht), polymorphic bands, and Shannon index were found in this population.
Conclusion
Through the study of partial diallels, we observed that horizontal and vertical resistance occur together in this patho-system. There is differential interaction between the genotypes and the isolates, and, based on the isolates’ reaction in the inoculated genotypes, it is possible to group them into physiological races. And it is also possible to observe that the genetic differences found do not constitute distinct pathogenic differences, because the two species cannot be differentiated based on the levels of aggressiveness with the genotypes of dry bean used in this study.
Thus, this study can contribute to understanding the populational structure of these two pathogens that are important for dry bean, about genetic and pathogenic diversity, as well as to understanding the interaction with in the host. Additionally, it can also contribute to dry bean breeding programs, in seeking resistance to common bacterial blight, about the development of lasting and broad-spectrum cultivars.
Acknowledgment
We acknowledge the Embrapa Arroz e Feijão, Universidade Federal de Goiás and Fapeg for providing financial support for this research, and CAPES, for financial support to B.A.R. de Paiva.
About the Authors
Corresponding Author
Wendland A
Programa de Pós Graduação em Genética e Melhoramento de Plantas, Universidade Federal de Goiás, Goiânia, Goiás, Brazil
- Email:
- adriane.wendland@embrapa.br
References
- Alves A, Henriques I, Santos A, Tacão M, Correia A (2003). Tipagem genética em microrganismos. MS in Molecular Microbiology. University of Aveiro, Aveiro, Portugal.
- Audy P, Laroche A, Saindon G, Huang HC, et al. (1994). Detection of the bean common blight bacteria, Xanthomonas campestris pv. phaseoli and X. c. phaseoli var. fuscans, using the polymerase chain reaction. Phytopathology. 84(10): 1185-1192. https://doi.org/10.1094/phyto-84-1185
- Kimati H, Amorim L, Rezende J, Bergamim Filho A, et al. (2005). Manual de fitopatologia: Volume 2: doenças das plantas cultivadas. (3thedn), Agronômia Ceres. São Paulo, SP.
- Birch PR, Hyman LJ, Taylor R, Opio AF, et al. (1997). RAPD PCR-based differentiation of Xanthomonas campestris pv. phaseoli and Xanthomonas campestris pv. phaseoli var. fuscans. European Journal of Plant Pathology. 103(9): 809-814.
- Boureau T, Kerkoud M, Chhel F, Hunault G, et al. (2013). A multiplex-PCR assay for identification of the quarantine plant pathogen Xanthomonas axonopodis pv. phaseoli. Journal of Microbiological Methods. 92(1): 42-50. https://doi.org/10.1016/j.mimet.2012.10.012
- Buiate EAS, Souza ED, Vaillancourt L, Resende I, et al. (2010). Evaluation of resistance in sorghum genotypes to the causal agent of anthracnose. Crop Breeding and applied biotechnology. 10(2): 166-172. https://doi.org/10.12702/1984-7033.v10n02a10
- Burkholder WH (1930). The bacterial diseases of the Bean. A comparative study. Memoir. Cornell Agricultural Experiment Station, 127.
- Coyne D, Schuster M (1974). Breeding and genetic studies of tolerance to several beans (Phaseolus vulgaris L.) bacterial pathogens. Euphytica. 23(3):651-656. https://doi.org/10.1007/bf00022486
- Davide LMC, Souza E (2009). Pathogenic variability within race 65 of Colletotrichum lindemuthianum and its implications for common bean breeding. Crop breed applied biotechnology. 9:23-30. https://doi.org/10.12702/1984-7033.v09n01a04
- De Bruijn FJ (1992). Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Applied and environmental microbiology. 58(7): 2180-2187.
- Duncan RW, Singh SP, Gilbertson RL (2011). Interaction of common bacterial blight bacteria with disease resistance quantitative trait loci in common bean. Phytopathology. 101(4):425-435. https://doi.org/10.1094/phyto-03-10-0095
- Excoffier L, Laval G, Schneider S (2005). Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary bioinformatics online, 1:47. https://doi.org/10.1177/117693430500100003
- Griffing B (1956). Concept of general and specific combining ability in relation to diallel crossing systems. Australian journal of biological sciences. 9(4): 463-493. https://doi.org/10.1071/bi9560463
- Goodwin P and Sopher C (1994). Brown pigmentation of Xanthomonas campestris pv. phaseoli associated with homogentisic acid. Canadian journal of microbiology. 40(1): 28-3. https://doi.org/10.1139/m94-005
- Hajri A, Brin C, Hunault G, Lardeux F, et al. (2009). A “repertoire for repertoire” hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One. 4(8):e6632. https://doi.org/10.1371/journal.pone.0006632
- Halfeld-Vieira BA, Souza RM, Figueira AR, Boari AJ (2001). Identification of Xanthomonas axonopodis pv. phaseoli and X. axonopodis pv. phaseoli var. fuscans through the PCR technique. Fitopatologia Brasileira. 26(4): 737-740. https://doi.org/10.1590/s0100-41582001000400008
- Hartl DL and Clark AG (2010). Princípios de genética de populações. Porto Alegre. Artmed, xii.
- López R, Asensio C, Gilbertson RL (2006). Phenotypic and genetic diversity in strains of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) in a secondary center of diversity of the common bean host suggests multiple introduction events. Phytopathology. 96(11): 1204-1213. https://doi.org/10.1094/phyto-96-1204
- Louws F, Fulbright D, Stephens CT, De Bruijn F (1995a). Differentiation of genomic structure by rep-PCR fingerprinting to rapidly classify Xanthomonas campestris pv. vesicatoria. Phytopathology. 85(5): 528-536. https://doi.org/10.1094/phyto-85-528
- Louws FJ, Fulbright DW, Stephens CT, de Bruijn FJ (1995b). Differentiation of Genomic Structure by rep-PCR Fingerprinting. Molecular plant pathology. 85: 528-536. https://doi.org/10.1094/phyto-85-528
- Mahuku GS, Jara C, Henriquez M, Castellanos G, et al. (2006). Genotypic Characterization of the Common Bean Bacterial Blight Pathogens, Xanthomonas axonopodis pv. phaseoli and Xanthomonas axonopodis pv. phaseoli var. fuscans by rep├ó┬?┬ÉPCR and PCR–RFLP of the Ribosomal Genes. Journal of phytopathology. 154(1): 35-44. https://doi.org/10.1111/j.1439-0434.2005.01057.x
- Manten J (2008). Importância de novos defensivos agrícolas no manejo de doenças no feijoeiro. In: Anais do Congresso Nacional de Pesquisa de Feijão, Campinas, SP, Brasil. p. 957-960.
- Manzanera AS, Carmen M, Asensio C, Singh SP (2006). Gamete selection for resistance to common and halo bacterial blights in dry bean intergene pool populations. Crop science, 46(1): 131-135. https://doi.org/10.2135/cropsci2005.0198
- Martin B, Humbert O, Camara M, Guenzi E, et al. (1992). A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Research. 20(13): 3479-3483. https://doi.org/10.1093/nar/20.13.3479
- Melo LC, Santos JB (1999). Identification of resistant genotypes considering polygenic systems in host-pathogen interaction. Genetics and Molecular Biology. 22(4): 601-608. https://doi.org/10.1590/s1415-47571999000400022
- Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, et al. (2013). Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS One, 8(3). https://doi.org/10.1371/journal.pone.0058474
- Mkandawire AB, Mabagala RB, Guzmán P, Gepts P, et al. (2004). Genetic diversity and pathogenic variation of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) suggests pathogen coevolution with the common bean. Phytopatholog. 94(6): 593-603.
- Munhoz C, Weiss B, Hanai L, Zucchi M, et al. (2011). Genetic diversity and a PCR-based method for Xanthomonas axonopodis detection in passion fruit. Phytopathology. 101(4): 416-424. https://doi.org/10.1094/phyto-06-10-0169
- Mutlu N, Vidaver A, Coyne D, Steadman J, et al. (2008). Differential pathogenicity of Xanthomonas campestris pv. phaseoli and X. fuscans subsp. fuscans strains on bean genotypes with common blight resistance. Plant Disease, 92(4): 546-554.
- Nei M (1973). Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences. 70(12): 3321-3323. https://doi.org/10.1073/pnas.70.12.3321
- Opio A, Allen DJ, Teri J (1996). Pathogenic variation in Xanthomonas campestris pv. phaseoli, the causal agent of common bacterial blight in Phaseolus beans. Plant pathology, 45(6): 1126-1133. https://doi.org/10.1046/j.1365-3059.1996.d01-187.x
- Parlevliet JE (1981). Disease resistance in plants and its consequences for plant breeding. In: Plant breeding II (Frey, KJ, ed). Iowa State University Press, Ames, 309-247.
- Pereira R, Souza E, Barcelos Q, Abreu A, et al. (2015). Aggressiveness of Pseudocercospora griseola strains in common bean genotypes and implications for genetic improvement. Genetic and Molecular Research. 14: 5044-5053. https://doi.org/10.4238/2015.may.12.7
- Rava CA (1984). Patogenicidade de isolados de Xanthomonas campestris pv. phaseoli. Pesquisa Agropecuária Brasileira, Brasília, 19:445-448.
- Rava C, Zimmermann M, Rocha M, Yamada T (1988). Crestamento bacteriano comum. In: Cultura do Feijoeiro: fatores que afetam a produtividade (Rava C, Sartorato A, eds.) Brasília, DF. Embrapa. 217-242.
- Restrepo S, Vélez CM, Verdier V (2000). Measuring the genetic diversity of Xanthomonas axonopodis pv. manihotis within different fields in Colombia. Phytopathology. 90(7): 683-690.
- Romeiro RS (2011). Bactérias fitopatogênicas. 2nd Ed. Viçosa, MG. Editora UFV.
- Santos AS, Bressan-Smith RE, Pereira MG, Rodrigues R, et al. (2003). Genetic linkage map of Phaseolus vulgaris and identification of QTLs responsible for resistance to Xanthomonas axonopodis pv. phaseoli. Fitopatologia Brasileira. 28(1): 5-10. https://doi.org/10.1590/s0100-41582003000100001
- Silva LL, Wendland A, Melo LC, da Costa JGC, et al. (2008). Interação diferencial entre isolados de Xanthomonas axonopodis pv. phaseoli e genótipos de feijoeiro. Documentos IAC, 85.
- Silva SAG, de Morais OP, Rava CA, da Costa JGC (2000) Método generalizado de análise de dialelos desbalanceados. Pesquisa Agropecuária Brasileira. 35(10): 1999-2005. https://doi.org/10.1590/s0100-204x2000001000011
- Yeh F, Boyle T, Xiyan JP (1999). Version 131: Microsoft Window-based freeware for population genetic analysis. University of Alberta and Center for International Forestry Research.
- Young J, Saddler G, Takikawa Y, De Boer S, et al. (1996). Names of plant pathogenic bacteria. 1864-1995. Review of Plant Pathology. 75(9): 721-763.
Keywords:
Download:
Full PDF- Share This