Mitochondrial diversity in the populations of Pediculus humanus capitis across eastern south of Tehran Province, Iran
Received: September 02, 2017
Accepted: September 28, 2017
Published: October 05, 2017
Genet.Mol.Res. 16(4): gmr16039826
DOI: 10.4238/gmr16039826
Abstract
Pediculus humanus capitis is a major obligate and district ecto-parasite of human which is distributed in different parts of the world and is under several research projects for population management and molecular analysis. In this study, one hundred and seventy head lice were collected from school girls at different high schools in five cities in south east of Tehran Province (Iran). They were analyzed using molecular methods for genotyping lice, Cytochrome Oxidase I (COI), for the first time in Iran. The phylogenetic analysis of the concentrated sequences of the head lice populations showed high divergence among the population collected from different cities. However, samples belonged to each city showed high homology with some of the GenBank sequences.
Introduction
Head lice, Pediculus humanus capitis De Geer 1767, is an obligatory blood-feeding ectoparasite of humans belonging to Pediculidae which aggregate and feed on the head region of humans, where females attach eggs to the base of hair shafts, are common amongst school-aged children, symptoms including itching and insomnia pose relatively few health risks (Light et al., 2008). Its effects as a serious pest and disease vector justify understanding the variation among different population of head lice in varied part of the world. Variation exists at all levels of biological organization, between and within species, populations and individuals of this pest (Olds, 2013). Molecular techniques have been used for insect species identification and were applied for the biological, evolutionary, phylogenic, and ecological studies (Sunantaraporn et al., 2015). It has been clear that one hundred and eight million base pairs (Mbp) genome of the body louse, Pediculus humanus corporis L., consisting of 10,775 genes were sequenced (Kirkness et al. 2010). On the other hand, the head louse, Pediculus humanus capitis, has not yet had its genome mapped, but previous work suggests these two organisms divergedfrom one another approximately 70,000 years ago with the adventure of clothing in humans (Kittler et al., 2003).
The current taxonomic position of human lice is unclear, as their description and phylogenetic classification have mostly been based on morphological criteria (Yong et al., 2003). To date, few genes or intergenic sequences have been studied for sucking lice: cytochrome oxidase I (COI) (Leo et al., 2002), cytochrome b (Cytb) (Boutellis et al., 2015; Page et al., 1998), elongation factor-1α (EF-1α) (Cruickshank et al., 2001), 12S rRNA (Page et al., 2002), 18S rRNA (Johnson et al., 2002), and two internal transcribed sequences (ITS1 and 2) (Leo and Barker, 2002). It should be noticed that collected data regarding diversity and distribution are largely based on studies of sequence variation in two mitochondrial genes, COI and cytb. Likewise, among these genes, mitochondrial genes have been employed to identify lineages and to analyze patterns of genetic diversity in P. h. capitis, especially COI have demonstrated to be one of the valuable phylogenetic tools for Phthiraptera (Yong et al., 2003). Kittler et al. (2003) investigated molecular evolution in P. humanus capitis with both nuclear (EF-1α,RPII) and mitochondrial (ND4, cytb) genes. Likewise, Light et al. (2008) investigated the taxonomic status of head lice using the mitochondrial (COI, cytb, ND4) loci. According to Cognato (2006), COI has been used in 84% of the genetic studies on different orders of Insecta. Mitochondria of P. h. capitis consists of 18 minicircles (Perlman et al., 2015). Head lice are divided into three clades A, B and C which are distributed geographically throughout the world. Boutellis et al. (2015) studied Cytb and MST and found different clades of head lice in Algeria for the first time. The patterns of genetic diversity in Asian and African head lice have seen much less study The current study begins to address this gap by examining sequence diversity in COI for different populations of head lice in Tehran province, Iran, to find any relationships in different populations from rural areas which are under the pest infection.
Material and Methods
One hundred and forty-three adult human lice from five rural areas named Varamin, Pakdasht, Qarchak, Pishva and JavadAbad in eastern south of Tehran Province origins were included in this study. Lice were collected from different infested school girls by local physicians. Samples were identified according to Kakrasulemankhel (2010). Lice were conserved in 95% ethanol at −20°C for further analysis.
The head lice specimens were removed from 95% ethanol by washing with proteinase K buffer, and then each sample was homogenized in 180 μl of lysis buffer LB and 20 μl of proteinase K. The genomic DNA was extracted using a rapid genomic DNA Isolation kit, fast and simple MBST (research Institute, Tehran, Iran) following the manufacturer’s instructions. The extracted head lice DNA was eluted in 50 μl of elution buffer, and the concentration was measured using Nano drop 2000c (Thermo-scientific, USA). The genomic DNA was stored for an extended time at −20°C until the next stage of the investigation. PCR amplification and sequencing were subsequently performed at the MBST, following standard protocol. Amplification of the COI employed the primer pair as below:
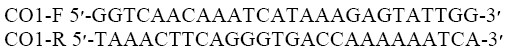
The following PCR Master Mix (CinnaGen, Tehran, Iran) were applied to prepare 50 μl DNA extractions: PCR buffer, 10 mM MgCl2, 0.4 mM F-Primer, 0.4 μM R-Primer, 0.2 μM, dNTPs, 200 mM, Taq, 0.5-2.5 units and DNA, 5 μl. The following conditions were applied: 95°C (1 min); 5 cycles of 95°C (45 sec), 50°C (40 s), 72°C (1 min) and final extension of 72°C (7 min), 37 cycles were applied for 5 cycles of 95°C (45 sec), 50°C (40 s), 72°C (1 min). 5 μl from each elute was analyzed on 1% agarose gel. The extracted DNA was amplified in PCR using primers derived from ß-actin. The forward and reverse sequences were assembled, edited and aligned using BioNeer (BioNeer, Corporation, North Korea) and sequenced by ABI PRISM Genetic Analyzer and then translated in MEGA (V6)50 to verify that they were free of stop codons and gaps and the phylogenetic tree was prepared by Bayesian method (Sunantaraporn et al. 2015).
Phylogenetic tree was constructed by MEGA 6.06 through neighbor joining. Then, all sequences generated in this study were deposited in GenBank under accession numbers as below:
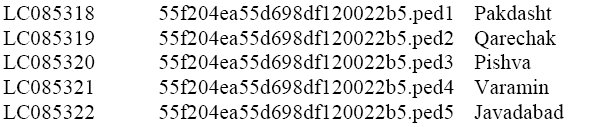
COI sequences obtained in this study were combined with those for head lice from GenBank to generate a global dataset to examine populations diversity in P. h. capitis in the studied area and again the phylogenetic tree was reconstructed to find out the similarity among the samples obtained in this study and the previous records on GenBank.
Results
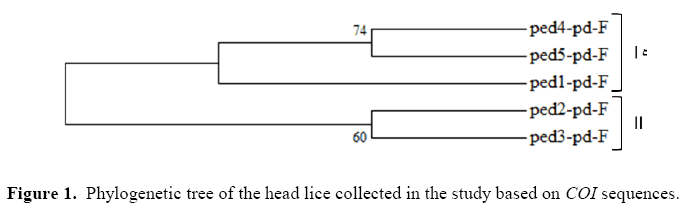
Molecular data from the mitochondrial COI gene were collected from 170 head lice and 560 base pairs were used for molecular analysis. For recognizing the phylogenetic relationship among the study samples, phylogenetic tree was constructed by MEGA 6.06 through neighbor joining and Clustalw algorithm. The results are represented in Figure 1. As it can be seen, the populations of five locations belonged to three distinct groups. The first group combined samples obtained from Varamin, Javad Abd by the bootstrap of 74. The second group consisted of populations belonging to Pishva and Qarchak with bootstrap of 60. And samples from Pakdasht showed low lineage to the other collected samples. As it can be seen, although the collected samples belonged to the same species, but some diversity in COI sequencing led them to different categories by varied similarities.
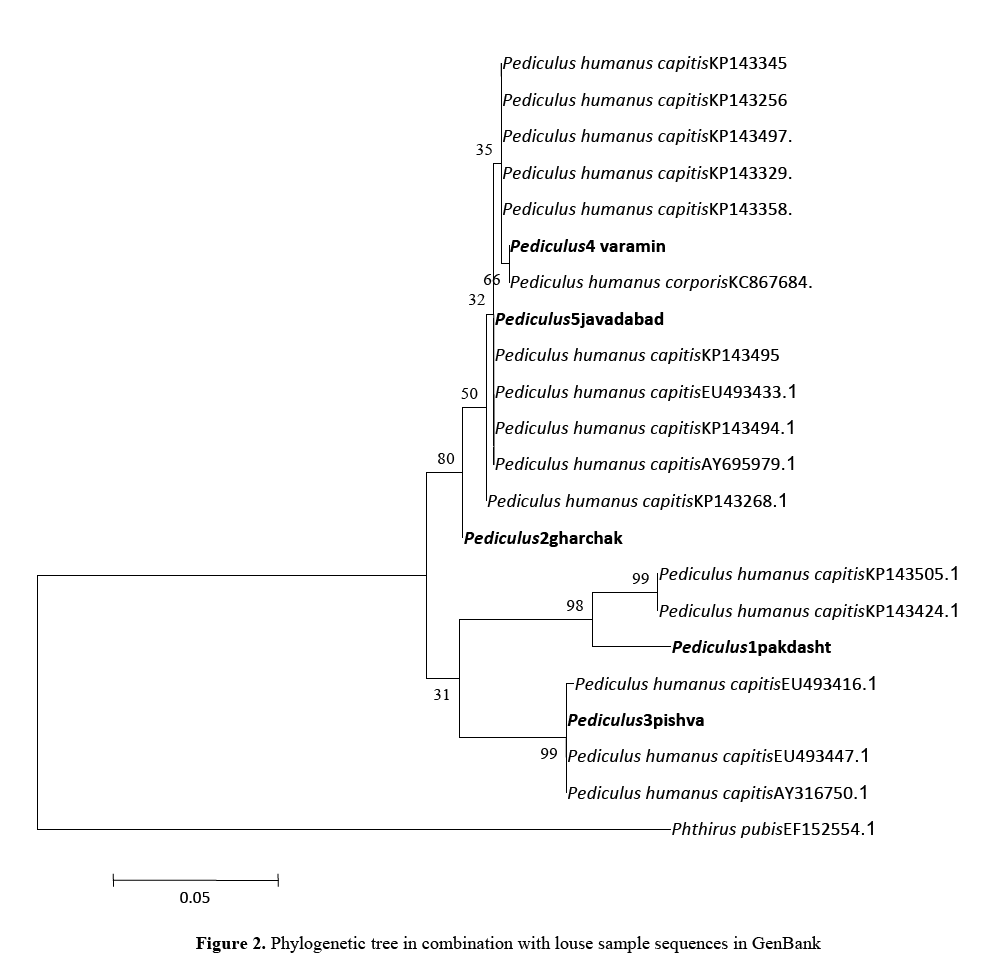
Again, phylogenetic tree was reconstructed in combination with louse sample sequences available in GenBank (Figure 2). As it can be seen, all the collected samples were belonged to the known species Pediculus humanus capitis, but there were some differences in COI sequences among the samples from different locations which ledto different bootstrap and lineage with samples recorded in GenBank from other part of the world. And the results in the phylogenetic trees (Figure 2) seems more complicated than the first grouping of the head lice in the study. As it can be seen mtDNA COI coding of samples from Varamin showed lineage with one record of P. humanus corporis (accession number: KC867684, Australia). However, it showed lineage with P. humanus capitis COI gene from Thailand (accession numbers: KP143345, KP 143256, KP133497, KP143329 andKP143358), too. Samples from Javadabad showed close similarity with the United States (accession number: EU493433, clade A) and the United Kingdom (Accession number: AY695979) and as an out group sample showed similarities with Thailand samples (accession numbers: KP143495, KP 143494 and KP143268). Again samples from Pakdasht showed lineage with the records from Thailand (accession numbers: KP143505 and KP143424) with bootstrap of 98. Finally samples from Pishva confirmed very close lineage with the records from different parts of the United States (accession number: EU493416, clade B; EU493447, clade B) and the United Kingdom (Accession number: AY316750).
Discussion
The genus Pediculus is currently recognized as including just one species in two sister isolates, P. humanus capitis and P. humanus corporis in Iran. Regional previous studies have been focused on the species distributionthroughout the country and this study can be considered as the one of the premium genomic identification report of Pediculu humanus from Iran. Kittler et al. (2003) studied genetic divergence between head and body lice distribution based on different nucleotide sequences including COI in 40 head lice from 12 countries which included Iran, too. Their study showed that in some countries such as Iran, Germany, Ethiopia, Philippine, Laos and Ecuador, there was high similarities between P. humanus capitis and Pediculus humanus corporis; so it was concluded that the topology of their phylogenetic tree indicated that body lice originated from head lice. This conclusion is justified by the results of this paper which showed sampled from Varamin had similarities and same origin with body lice. Leo and Barker (2005) isolated DNA from one head loos from Iran and their study showed the divergence of head and body lice in different countries including Iran. The results of this study showed high diversity among the samples collected from five cities of Tehran province according to COI sequences. Previously Ashfaq et al. (2015) demonstrated that there is 10% probability of diversity in COI sequence among the head lice samples collected from different part of the world.
Human lice have been classified based on mitochondrial DNA into three phylogenetic clades (Raoult et al. 2008). Veracx et al. (2013) by studying different population of head lice in Africa belonging to the three clades of A, B and C found out that clades A and B are sisters and the differences between them could not be significantly important. The same study insisted on the relevancy of Clades A and B in North America. Boutellis et al. (2013) reported that each clade has a unique geographical distribution: Clade A can be found worldwide,while clade B can be found in North America, Central America, Europe, Australia (Raoult et al. 2008) and Algeria (Boutellis et al., 2015), and clade C can be found in Nepal (Reed et al. 2004), Ethiopia (Angelakis et al., 2011) and Senegal (Boutellis et al., 2012). Globalization could be the main reason for the spread distribution of head lice in all parts of the world (Boutellis et al., 2013). The presence of deeply divergent mitochondrial clades is possibly the result of retained ancestral polymorphism, multiple colonization events of lice on their modern human hosts from now extinct archaic hominids (Creer et al., 2001; Reed et al., 2004), and/or multiple modern human and parasite migration events (Arau´jo et al., 2008), or a combination. Ashfaq et al. (2015) added two new clades, D and E to the previous classifications according to their origin which totally have maximum divergences of 10.3% at COI. and. Ashfaq et al. (2015) showed that previous clade classification of head lice throughout the world is not recently supported and early human migration patterns does not justifies the present distribution of Pediculus humanus caldes, as they reported clade B from new origin of Africa and their interpretation justifies our results.
The phylogeny shows that Pediculus species on chimpanzees and humans are sisters, which form a clade with Pthirus that is sister to Pedicinus, the most basal member of Phthiraptera. Bootstrap support for theserelationships is high (Boutellis et al., 2015; Reed et al., 2004) which can justifies the high bootstrap of the collected samples from Varamin with the GeneBank recorded data for the body louse, P. humanus corporis. Recently, it was concluded that human head and body lice are two ecotypes of lice that have identical genomic content but different in gene expression patterns that result in different phenotypes (Veracx and Raoult 2012); which would explain the result obtained for samples from Varamin. Reed (2004) reported that two clades of head lice are more common than the third one and these two clades have the same recent ancestor 0.54 million years ago. According to Li et al. (2010), phylogenetic organizations of head and body lice based on each of the four intergenic spacers which led to 96 genotypes in 97 samples of head lice support the hypothesis that head lice were grouped with body lice in the same clusters or sub-clusters which supports the high diversity of COI sequencing in the present study
Conclusion
The main aim of this study was to identify COI sequences in Pediculus humanus capitis from samples collected from different cities in Tehran which has not been studies previously and this report could be considered as the first regional study based in Iran on COI diversity in head lice. As it can be seen in the phylogenetic tree, the origin of the collected samples showed high diversity. On the other hand, although clade identification was not the aim of the study, surprisingly the obtained results showed the clade similarities of the collected samples with the recorded sequences in GenBank which led us to investigate further studies based on the clade identification of the collected samples. Although the present results admitted the effect of human geographical migration affect on the distribution of the head lice clades. Additional studies on other mitochondrial genes especially Cytb are needed to confirm the obtained data in this paper and also to admit the presence of head lice clade B in Iran.
About the Authors
Corresponding Author
Neda Kheradpir
Department of Entomology, Fac. Agriculture, Islamic Azad University, Varamin (Pishva) Branch, Tehran, Iran
- Email:
- Kheradpir@iauvaramin.ac.ir
References
- Angelakis E, Diatta G, Abdissa A, Trape JF, et al. (2011). Altitude-dependent Bartonella Quintana genotype C in head lice, Ethiopia. Emerg. Infect. Diseases. 17: 2357–2359.https://doi.org/10.3201/eid1712.110453
- Arau Jo A, Ferreira LF, Maues de Serra Freire N, Reinhard KJ, et al. (2008). Ten thousand years of head lice infection. Parasitol. Today, 16:269 https://doi.org/10.1016/s0169-4758(00)01694-x
- Ashfaq M, Prosser S, Nasir S, Masood M, et al. (2015). High diversity and rapid diversification in the head louse, Pediculus humanus (pediculidae: Phthiraptera). Sci. Reports. 5:14188. https://doi.org/10.1038/srep14188
- Boutellis A, Bitam I, Fekir K, Mana N (2015). Evidence that clade A and clade B head lice live in sympatry and recombine in Algeria. Med. Vet. Entomol. 29: 94-98.https://doi.org/10.1111/mve.12058
- Boutellis A, Drali R, Rivera M, Mumcuoglu K, et al. (2013) Evidence of sympatry of clade A and clade B head lice in a pre-Columbian Chilean mummy from Camarones. PLoS One, 8, e76818. https://doi.org/10.1371/journal.pone.0076818
- Cognato AI (2006). Standard percent DNA sequence difference for insects does not predict species boundaries. J. Econ. Entomol. 99(4): 1037-1045. https://doi.org/10.1603/0022-0493-99.4.1037
- Creer S, Malhotra A, Thorpe RS, Chou WH. (2001). Multiple causation of phylogeographical patterns as revealed by nested clade analysis of the bamboo viper (Trimeresurus stejnegeri) within Taiwan. Mol. Ecol. 10: 1967-1981. https://doi.org/10.1046/j.0962-1083.2001.01332.x
- Cruickshank RH, Johnson KP, Smith VS, Adams RJ, et al. (2001). Phylogenetic analysis of partial sequences of elongation factor identifies major groups of lice (Insecta: Phthiraptera). Mol. Phylogenet. Evol. 19(2): 202-215. https://doi.org/10.1006/mpev.2001.1033
- Johnson KP, Allen JM, Olds BP, Mugisha L, et al. (2014). Rates of genomic divergence in humans, chimpanzees and their lice. Proc. R. Society of B. 281.https://doi.org/10.1098/rspb.2013.2174
- Johnson KP, Williams BL, Drown DM, Adams RJ, et al. (2002). The population genetics of host specificity: Genetic differentiation in dove lice (Insecta: Phthiraptera). Mol. Ecol.11:25-38 https://doi.org/10.1046/j.0962-1083.2001.01412.x
- Kakarsulemankhel JK (2010). Re-description of Pediculus humanus corporis Linnaeus, 1758 (Anoplura). Pakistan J. Entomol. Karachi. 25 (2): 101-106.
- Kirkness EF, Haas BJ, Sun W, Braig HR, et al. (2010). Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Nat. Acad. Sci. United States of America (PNAS), 107(27): 12168-12173.
- Kittler R, Kaysar M, Stoneking M (2003). Molecular evolution of Pediculus humanus and the origin of clothing. Curr. Biol. 14: 2309. https://doi.org/10.1016/j.cub.2004.12.024
- Leo N P and Barker SC. (2005). Unravelling the evolution of the head lice and body lice of humans. Parasitol. Res. 98: 44-47. https://doi.org/10.1007/s00436-005-0013-y
- https://doi.org/10.3201/eid1712.110453
- Leo NP, Campbell NJ, Yang X, Mumcuoglu K, et al. (2002). Evidence from mitochondrial DNA that head lice and body lice of humans (Phthiraptera: Pediculidae) are conspecific. J. Med. Entomol. 39(4): 662-666. https://doi.org/10.1603/0022-2585-39.4.662
- Li W, Ortiz G, Fournier PE, Gimenez G, et al. (2010). Genotyping of human lice suggests multiple emergences of body lice from local head louse populations. PLoS Negl Trop Dis. 4(3): e641. https://doi.org/10.1371/journal.pntd.0000641
- Light JE, Allen JM, Long LM, Carter TM, et al. (2008). Geographic distributions and origins of human head lice (Pediculus humanus capitis) based on mitochondrial data. J. Parasit. 94: 1275-1281.https://doi.org/10.1645/ge-1618.1
- Olds B (2013). Analysis of genetic variation in Pediculus humanus and Populus trichocarpa. PhD Dissertation, University of Illionis at Urbana-Champaign,University of Illionis at Urbana-Champaign
- Page RD, Lee PL, Becher SA, Griffiths R, et al. (1998). A different tempo of mitochondrial DNA evolution in birds and their parasitic lice. Mol. Phylogenet. Evol. 9: 276–293.https://doi.org/10.1006/mpev.1997.0458
- Perlman ST, Hodson CN, Hamilton PT, Opit GP, et al. (2015). Maternal transmission, sex ratio distortion, and mitochondria. PNAS. 112 (33): 10162-10168. https://doi.org/10.1073/pnas.1421391112
- Raoult D, Reed DL, Dittmar K, Kirchman J, et al. (2008). Molecular identification of lice from pre-Columbian mummies. J. Infectious Dis. 197(5): 35-43.
- Reed DL, Smith VS, Hammond SL, Rogers AR (2004). Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2(11): 1972-1983. https://doi.org/10.1371/journal.pbio.0020340
- Sunantaraporn S, Sanprasert V, Pengsaku T, Phumee A, et al. (2015). Molecular survey of the head louse Pediculus humanus capitis in Thailand and its potential role for transmitting Acinetobacter spp. Parasit Vectors. 8 (127): 1-7. https://doi.org/10.1186/s13071-015-0742-4
- Veracx A, Boutellis A, Raould D (2013). Genetic recombination events between sympatric Clade A and Clade C in Africa. J. Med. Entomol. 50(5): 1165-1168. https://doi.org/10.1603/me13028
- https://doi.org/10.1016/s0169-4758(00)01694-x
- Mitochondrial diversity in the populations of Pediculus humanus capitis across eastern south of Tehran Province, Iran
- Yong Z, Fournier PE, Rydkina E, Raoult D. (2003). The geographical segregation of human lice preceded that of Pediculus humanus capitis and Pediculus humanus humanus. C. R. Biol. 326: 565-574. https://doi.org/10.1016/s1631-0691(03)00153-7
Keywords:
Download:
Full PDF- Share This