MicroRNAs as biomarkers of kidney allograft injuries ischemia reperfusion and acute rejection
Received: August 21, 2018
Accepted: October 25, 2018
Published: November 05, 2018
Genet.Mol.Res. 17(4): http://dx.doi.org/gmr16039933
DOI: http://dx.doi.org/10.4238/gmr16039933
Abstract
Renal transplantation provides a significant increase in life expectancy for patients with end-stage kidney diseases. Despite the unequivocal progresses made in the last decades, many grafts injuries still jeopardize graft function leading to its loss. Currently, the allograft biopsy is the gold standard for diagnosing such conditions. However, it has many limitations including risk and cost. Therefore, it is desirable to develop non-invasive biomarkers able to lead to the diagnosis of graft injuries. This is particularly true during the delayed graft function (DGF) period in which the functional parameters are not available. MicroRNAs regulate their target genes through mechanisms of translational repression and messenger RNA (mRNA) degradation. A single miRNA can regulate the expression of hundreds of mRNAs and proteins, deregulation of miRNAs can lead to disruption and suppression of genes that operate in intracellular signaling cascades leading to disease conditions. Technological advances have allowed the accurate detection of miRNAs in biological fluids providing either qualitative or quantitative results. Here we reviewed the literature in which miRNAs were analyzed as injury markers in renal transplantation mainly for contribute the diagnosis of acute rejection (AR). In cross sectional studies miRNAs were found to be useful markers of AR in renal transplant patients and their heightened expression by may became useful on aiding the diagnosis of AR during the DGF period.
Introduction
Kidney transplantation has become the treatment of choice for many patients with end-stage renal failure. Compared to other forms of renal replacement therapy, transplantation provides a significant increase in life expectancy and quality of life. Acute rejection (AR) is a major complication following renal transplantation and may lead to its failure at any time during the post-transplant course. Under the currently employed immunosuppressive regiments, AR occurs in around 10-20% of patients during the first post-transplant year. Besides rejection many other injuries may jeopardize grafts function and survival, including mainly infections (fungal, bacterial and viral) and drug toxicities. For most situations the diagnosis relies on a biopsy that is performed to elucidate graft dysfunction and currently there are no reliable non-invasive tools to the diagnosis prior to the occurrence of graft dysfunction. Overtime non-invasive molecular diagnosis of allograft rejection have being developed by using body fluids, such as urine and peripheral blood, for mRNA and microRNAs evaluation as candidate biomarkers. The objective of this article is to review the current status of the studies involving microRNAs as biomarkers of kidney allograft injuries specially ischemia-reperfusion and AR.
Literature Review
Discovery and biogenesis of microRNAs
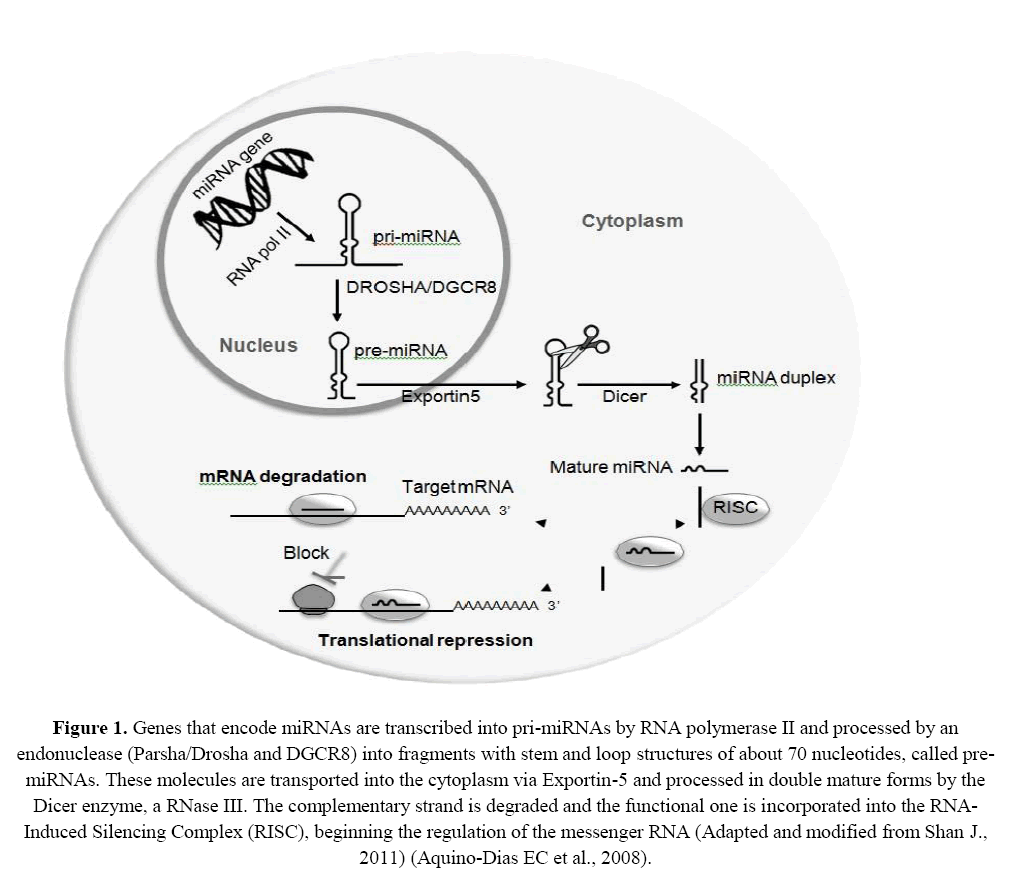
MicroRNAs (miRNAs) are endogenous small non-coding RNAs of approximately 18-22 nucleotides in length, involved in gene regulation, that play key roles in many biological pathways in plants, animals and viruses (Bartel DP, 2009 ). These molecules are specifically encoded by genes that are mainly located in intergenic or intronic regions of their target genes (Alarrayed S, 2011). Since the discovery of miRNAs, in 1993, the knowledge on their biogenesis, mechanisms of action and biological functions has improved remarkably (Bartel DP, 2009; Chang TC and Mendell JT, 2007; Marí-Alexandre J et al., 2016; Reinhart BJ et al. 2000) miRNAs are transcribed by RNA polymerase II into primary transcripts called pri-miRNAs. These are long structures that have 5’ (a 5'-triphosphate bond between the 5' precursor molecule, a nucleotide for protection against ribonucleases) and a poly (A) tail that allows protein binding and ensure stability of the molecule. Pri-miRNAs are arranged in a central stem, in which two RNA segments with complementary bases (by Watson-Crick pairing) are paired [Bartel DP, 2004 ; Li JY, Yong TY, Michael MZ, Gleadle JM, 2010).
This sequence is flanked at one side by other non-paired bases and across the loop region, where the base pairs are not complementary, forming circular handles, assuming the format of a "hair clip" (hairpin). Inside the core, non-paired regions are cleaved by RNaseIIIDrosha and Parsha/DGCR8. As a result, only the stem and loop regions remain with approximately 70 nucleotides in length, called precursor miRNAs (pre-miRNAs) (Bartel DP, 2004; Sarma NJ et al., 2012). These pre-miRNAs are then transported to the cytoplasm via exportin-5 and cleaved by the RNase III Dicer enzyme complex/argonaut, which removes the terminal loop forming miRNAs-duplex of approximately 22 nucleotides with short 3'-overhangs. One strand of the miRNA is degraded by an ATP-dependent RNA helicase; the other strand, that remains functional, is incorporated into the RNA-induced silencing complex (RISC). RISC is composed of several proteins that guide the silencing machinery to the target messenger RNA (mRNA), inhibiting its transcription or promoting its degradation (Figure 1) (Bartel DP, 2004; Li JY, Yong TY, Michael MZ, Gleadle JM, 2010; Sarma NJ et al., 2012).
Figure 1: Genes that encode miRNAs are transcribed into pri-miRNAs by RNA polymerase II and processed by an endonuclease (Parsha/Drosha and DGCR8) into fragments with stem and loop structures of about 70 nucleotides, called pre-miRNAs. These molecules are transported into the cytoplasm via Exportin-5 and processed in double mature forms by the Dicer enzyme, a RNase III. The complementary strand is degraded and the functional one is incorporated into the RNA-Induced Silencing Complex (RISC), beginning the regulation of the messenger RNA (Adapted and modified from Shan J., 2011) (Aquino-Dias EC et al., 2008).
The miRNAs mechanisms of action involve their binding primarily to the 3'- untranslated region (UTR) of mRNA targets through position 2 to 8 nucleotides at the 5' end of miRNA' sequence, also known as seed sequence miRNAs regulate their target genes through two mechanisms, repression of translation or mRNA degradation. Perfect or near-perfect match between miRNA and its target mRNA target leads to mRNA degradation, while imperfect pairing mediates mRNA translational repression. However, there is still at lack of knowledge about the reasons why a specific miRNA choose one mechanism instead of the other one (Harris A, Krams SM, Martinez OM, 2010; Kim DH, Saetrom P, Snøve O Jr, Rossi JJ 2008; Shan Jet al., 2011).
A unique miRNA can control hundreds of different mRNAs and related biological pathways. At the same time, several miRNAs may regulate a specific mRNA (Nassirpour R, Raj D, Townsend R, Argyropoulos C, 2016). These small RNA molecules exert control of processes such as development, cell proliferation, differentiation, apoptosis, metabolism and oncogenesis (Bartel DP, 2009). The dysregulation of such processes can lead to disruption and suppression of the genes that operate in the intracellular cascade signaling, resulting in new diseases or progression of already existent diseases (Kim DH, 2008; Solez K et al., 2008 ;Kato M et al., 2009 ;Lee SO, et al., 2008; Li JY et al., 2010 ;Livak KJ and Schmittgen TD, 2001 ;Lorenzen JM, 2011 ;Lorenzen JM et al., 2011; Lorenzen JM, 2015; Lucile A et al., 2016; Meier-Kriesche HU et al. 2001; Melkonyan HS et al. 2008; Mas VR et al., 2013, Marí-Alexandre J et al., 2016; Newell KA et al., 2010;O’Connell RM et al., 2007; Grosso G et al., 2012 ; Reinhart BJ et al., 2000; Serón D et al., 1989; Suthanthiran M, 1998; Haas M et al., 2014; Sui Wet al., 2008; Shan J et al., 2011). Previous studies suggest that miRNAs actively participate in the development and regulation of immune cells. Specific miRNAs have been identified as having significant impact on the differentiation of B and T cells and on the cellular process required for innate and adaptive immunity, including inflammation, signaling through T cell receptors and "toll-like" cell receptors" (TLR), production of cytokines, cellular function of T regulatory cells and presentation (Aquino-Dias EC et al., 2008;Harris A, Krams SM, Martinez OM, 2010; Newell KA et al., 2010; O’Connell RM et al., 2007). Several diseases have been linked to disorders of the signaling process (Marí-Alexandre J et al., 2016).
Ischemia and reperfusion of kidney allografts
After organ retrieval, especially in deceased donor transplantation, the kidney goes through a period of cold ischemia time while it is preserved by cold static storage or pulsatile machine perfusion. During this period there is no blood circulation, which is only restored after vascular sutures and release of the surgical clamps (Livak KJ and Schmittgen TD, 2001). These processes lead to the ischemia and reperfusion injuries and trigger a non-specific inflammatory cascade, characterized by lymphocytic infiltration followed by injuries to the parenchymal tissue and causing damage to the graft (Alarrayed S et al., 2011; Solez K et al., 2008; GwinnerW 2007 ; Livak KJ and Schmittgen TD, 2001). Briefly, hypoxia due to low oxygen availability decreases adenylatecyclase activity, cyclic adenosine monophosphate (cAMP) levels and intracellular adenosine triphosphate (ATP) activity. Also, there is a dysregulation of calcium homeostasis that occurs concomitantly to an increment in vascular permeability (Geddes CC, Woo YM, Jardine AG, 2002; Newell KA et al., 2010). These processes lead to the activation of cell death programs such as apoptosis, necrosis and autophagy as a result of significant changes in gene transcription (Newell KA et al., 2010). During the DGF period the renal graft has poor or no function and this is mostly due to ischemia and reperfusion injury (IRI). This condition is associated with increased risk of allograft rejection, both acute and chronic, ultimately resulting in graft loss (Siedlecki A, Irish W, Brennan DC, 2011; Grosso G et al., 2012). DGF is the most common complication in the immediate post-transplant period and is usually defined by the need for dialysis in the first week after transplantation (Huang X, Le QT, Giaccia AJ, 2010).
As the blood flow to the graft is reestablished, right after the release of the vascular clamps, the reperfusion injuries start to take place. Reperfusion leads to the production of oxygen-free radicals and activates an inflammatory response (Lorenzen JM, 2015). IRI is a complex situation involving renal vasoconstriction and tubular damage. The proposed mechanisms include anoxia, release of reactive oxygen molecular species, especially superoxide, hydrogen peroxide and radical hydroxyl (Bijkerk R et al., 2014; Serón D et al., 1989). Neutrophils influx is followed by the release of oxygen free radicals (Girlanda R and Kirk AD, 2007; Meier-Kriesche HU et al., 2001) Calcium is a mediator of injury of failure participating in the pathogenesis of vasoconstriction and mesangial constriction (Bhatt K, Mi QS, Dong Z, 2011).
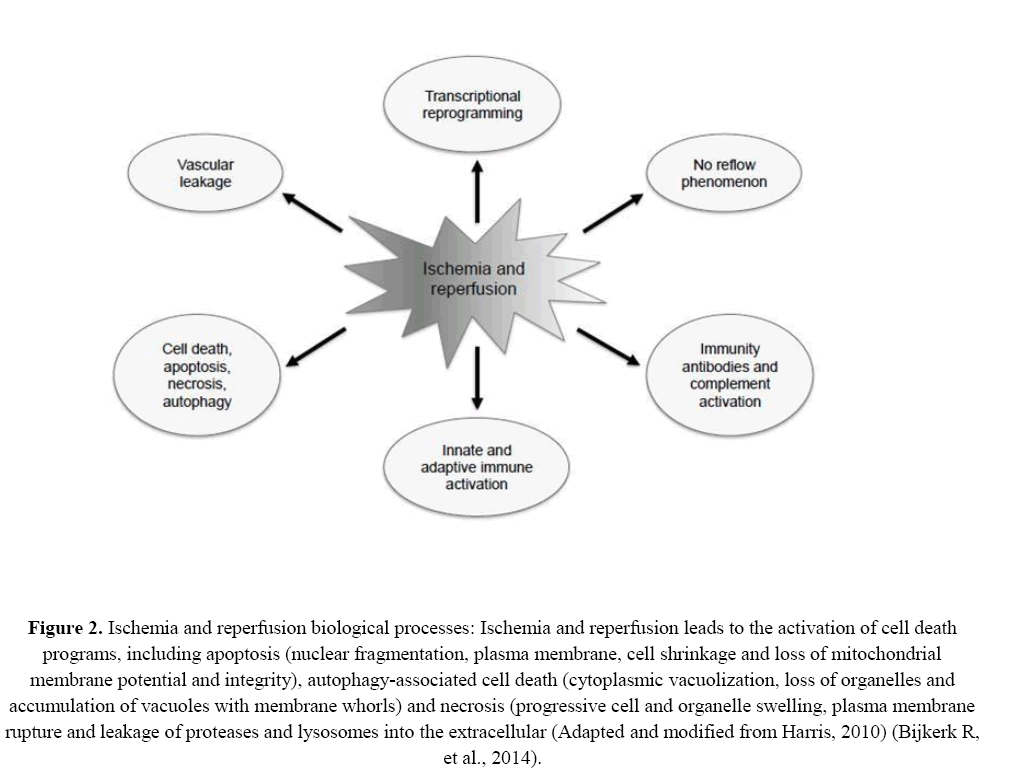
IRI injuries facilitate the activation of immune pathways, including the recognition of new antigens by antibodies and activation of the complement system (Lorenzen JM, 2015; Serón D, et al., 1989)These important and inevitable processes have the potential to affect cellular integrity and are major players in the initial injury to the graft. In the long term they are involved in the occurrence of structural changes, including interstitial fibrosis (Bhatt K, Mi QS, Dong Z, 2011). Figure 2 depicts schematically the steps involved in the ischemia and reperfusion injuries of organ grafts.
Figure 2: Ischemia and reperfusion biological processes: Ischemia and reperfusion leads to the activation of cell death programs, including apoptosis (nuclear fragmentation, plasma membrane, cell shrinkage and loss of mitochondrial membrane potential and integrity), autophagy-associated cell death (cytoplasmic vacuolization, loss of organelles and accumulation of vacuoles with membrane whorls) and necrosis (progressive cell and organelle swelling, plasma membrane rupture and leakage of proteases and lysosomes into the extracellular (Adapted and modified from Harris, 2010) (Bijkerk R, et al., 2014).
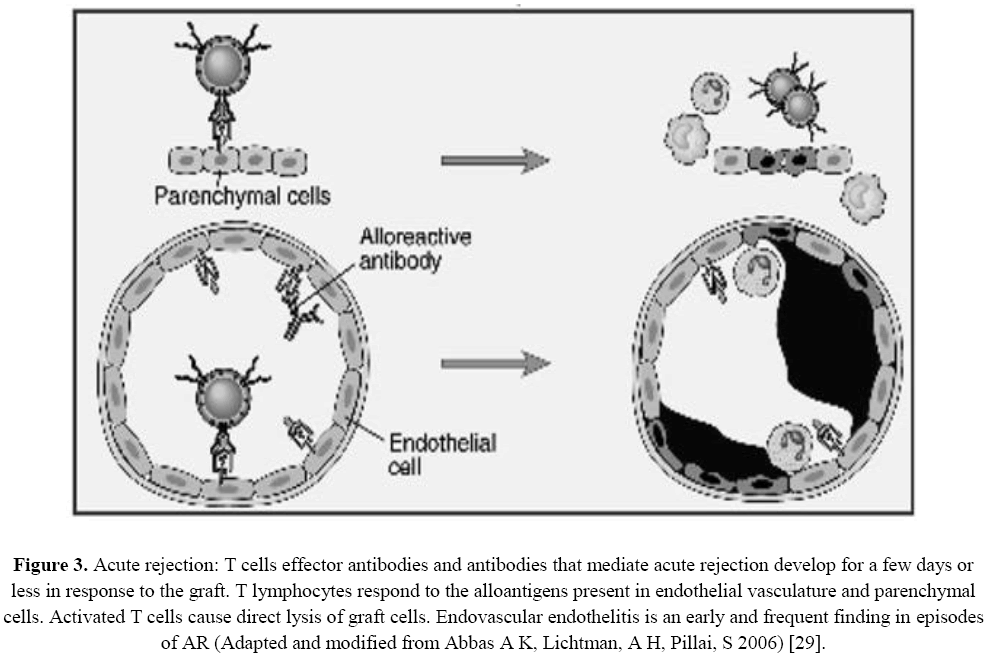
Figure 3: Acute rejection: T cells effector antibodies and antibodies that mediate acute rejection develop for a few days or less in response to the graft. T lymphocytes respond to the alloantigens present in endothelial vasculature and parenchymal cells. Activated T cells cause direct lysis of graft cells. Endovascular endothelitis is an early and frequent finding in episodes of AR (Adapted and modified from Abbas A K, Lichtman, A H, Pillai, S 2006) [29].
miRNAs as Biomarkers of Chronic Kidney disease ( CKD)
The miRNAs can be used as markers of chronic graft damage, have been shown to be involved in the progression of CKD and the development diabetes. CKD is a progressive loss of kidney function associated with persistent (> 3 months) glomerular filtration impairment and/or renal damage (Nassirpour R, Raj D, Townsend R, Argyropoulos C, 2016).
The potential of miRNA as biomarker of CKD is clinically important of chronic renal impairment: diabetic kidney disease (CKD) and chronic allograft nephropathy (CAN). Preclinical and clinical evidence suggests that miRNAs in biofluids (e.g. plasma and urine) may reflect both specific and non-specific effects of renal filtration and damage. Exploitation of non-specific effects may lead do the introduction of better filtration markers and renal damage markers for use in clinic or when dosing drugs with narrow therapeutic range in patients with CKD (Nassirpour R, Raj D, Townsend R, Argyropoulos C, 2016).
Biomarkers levels could be useful for risk evaluation and have a prognostic value in terms of treatment outcome, the microRNAs are novel response parameters and sensitivity predictors for cancer treatment. Chronic kidney disease-mineral bone disorder (CKD-MBD) is characterized by vessel and bone damage secondary to progressive loss of kidney function, miRNA could also be potential targets for innovative therapies, using the panel of ground-breaking tools now available for researchers and clinicians (Metzinger-Le Meuth V et al., 2017)
miRNAs signatures might help to assess pathophysiological changes during chronic kidney disease (CKD) and potential bone specific and vascular risks as well as therapy aspects in these patients. How example, the relative expression of miR-223-3p and miR-93-5p was down-regulated in patients with CKD stage 4 and 5 compared to healthy controls. MiR-223-3p and miR-93-5p were associated with interleukin-6 (IL-6) and eGFR levels, and by trend with interleukin-8 (IL-8), C-peptide, hematocrit, and parathyroid hormone (PTH). Identified miRNA signatures can contribute to future risk markers or future therapeutic targets in bone and kidney disease (Ulbing M et al., 2017)
Acute rejection of kidney allografts
Acute rejection (AR) is still a frequent and ominous complication in clinical kidney transplantation (Meier-Kriesche HU et al., 2001). It poses a risk for allograft loss, partial or complete permanent loss of function and it may occur at any time after transplantation. AR involves cellular and antibody-mediated components and may affect several organ structures initiating by graft endothelium (Girlanda R and Kirk AD, 2007).
This process of vascular and parenchymal lesion mediated by T-cells and antibodies, which usually begins after the first week of transplantation, T efector cells and antibodies that mediate acute rejection develop for a few days or less in response to the graft, T lymphocytes respond to the alloantigens present in the endothelial vasculature and parenchymal cells. Activated T cells cause direct graft cells endovascular endothelitis is an early and frequent finding in episodes of acute rejection (Abbas A K, Lichtman A H, Pillai S, 2006).
The major targets of the immune system are tubular epithelial cells and surrounding structures, glomerular capillaries and allograft vessels (Haas M et al., 2014). Acute cellular is characterized predominantly by T lymphocytes infiltrate accompanied by B lymphocytes, macrophages, and granulocytes (Lee SO et al., 2008; Melkonyan HS et al., 2008). Anti-HLA antibodies recognize specific sequences of amino acids present in the HLA antigens expressed on the surface of endothelial cells of the graft and activate the complement cascade, producing damage of the graft microcirculation (Eltzschig HK and Eckle T, 2011; Melkonyan HS et al., 2008).
Anti-allograft immunological reactions may exist sub-clinically and may also persist after a rejection treatment in both cases eliciting mechanisms that ultimately lead to progressive graft fibrosis followed by loss of function (Kato M et al., 2009). Rejections are usually clinically suspected on the scenario of graft dysfunction and currently graft biopsies interpreted according to the Banff 07 classification are the gold standard for the diagnosis (Anglicheau D et al., 2009). However, the biopsy approach has many limitations such as insufficient sampling (as a consequence of the focal nature of the rejection processes), poor reproducibility of interpretation, complications (bleeding, hematomas, arterio-venous fistulas, graft loss and even death) and high cost. Finally, biopsies accuracy cannot be validated due to the fact that there is no other independent methodology for assessing the existence of rejection (Eltzschig HK and Eckle T, 2011; Melkonyan HS et al., 2008).
These limitations have encouraged the search for alternative methods for the diagnosis of graft injuries including acute rejection. To date, among the non-invasive molecular methods that are under development, miRNAs analyzes are shading new light as promising biomarkers for the diagnosis of injuries associated to renal transplantation (Haas M et al., 2014).
As miRNAs play an important role in the regulation of protein synthesis, the assessment of their expression may provide information regarding organ physiology in normal and diseased states. In an expression atlas of human and mouse miRNAs, great similarity between species was found, and exclusive expression in a specific organ appeared to be very rare (Hartono C, Muthukumar T, Suthanthiran M, 2010; Melkonyan HS et al., 2008). Most miRNAs were found in more than one organ. Finally, expression of most miRNAs was ubiquitous and some showed some degree of tissue specificity (Hartono C, Muthukumar T, Suthanthiran M, 2010).
Conceptual map
Renal transplantation is the treatment of choice for many patients with chronic terminal loss of renal function since it offers a significant increase in patients' expectations and quality of life. As already mentioned and evidenced since the beginning of the practice of transplants between genetically distinct individuals, tissues and organs lose their functions through a process mediated by the immune system which is called rejection. Such a process is partially controlled by modifying the receptor's immune response with immunosuppressive drugs and biological agents.
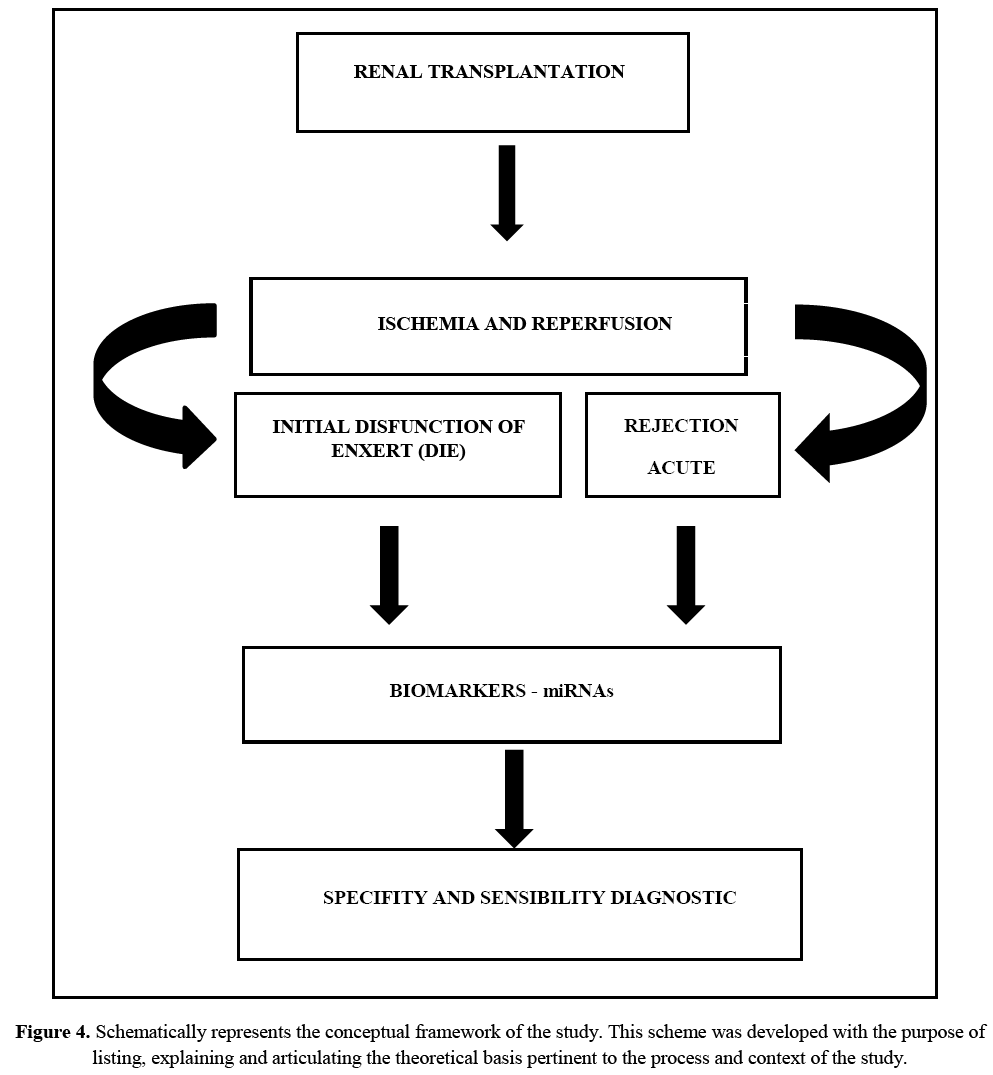
Injury to ischemia and reperfusion is an inevitable process in renal transplantation, with among its consequences a delay in graft function. These insults, which are responsible for initial graft dysfunction (DIE), by triggering or facilitating the mechanisms that favor acute rejection (AR) and by programming gene, metabolic and tissue changes that culminate in the graft fibrosis process and its chronic loss, the activation of the immune system, stimulating both the innate and the adaptive responses.
AR is an important risk factor for graft loss, being a frequent and serious post-transplant complication. Biopsy is costly and has problems related to representativeness and variability of interpretation. Nevertheless, in current practice, it is still the gold standard for the diagnosis of renal graft dysfunctions. Accurate non-invasive biomarkers that can affirm with high sensitivity and specificity the allograft status are necessary to improve the practice of transplants.
Figure 4 schematically represents the conceptual framework of the study. This scheme was developed with the purpose of listing, explaining and articulating the theoretical basis pertinent to the process and context of the study.
Molecular biomarkers studies involving miRNAs in renal transplantation
In the last two decades molecular biomarkers have been actively searched in the field of organ transplantation. Most of the research in this field has being performed in renal transplantation analyzing mRNA in biopsies, urine and peripheral blood lymphocytes utilizing either high-throughput platforms and/or real-time quantitative reverse transcription PCR (qRT-PCR), which allowed the monitoring of DNA amplification throughout the process.
The real-time PCR technique analyzes the gene expression, requiring the extraction of the genetic material (RNA) from the tissue of interest. Single-stranded RNA is unstable and needs to be synthesized in a complementary molecule (cDNA) through the action of a reverse transcriptase, using the enzyme Taq polymerase, deoxynucleotides, ions and primers, (specific DNA primer sequences of interest that will signal from which point the enzyme will start the sequence). The great majority of these studies are cross-sectional studies with few follow-up studies (Kato M et al., 2009; Lee SO et al., 2008; Haas M et al., 2014).
More recently miRNAs analyzes have been proposed for aiding diagnosis, establishing prognosis and evaluate response to the therapy in renal diseases and transplantation and are thought to provide promising biomarker candidates due to their stability and presence in biological fluids such as urine and blood (Hartono C, Muthukumar T, Suthanthiran M, 2010; Melkonyan HS et al., 2008).
Upon the discovery of miRNAS, some research groups have studied them as possible biomarkers of renal allograft injuries. In the review of the literature, the main aspects related to the factor under study with key words:“miRNAs”, “Rejection Acute”, “kidney Transplantation”, “Ischemia and Reperfusion”, “microRNAs”, “miRNAs” and “Kidney Transplantation”,“miRNAs” and “Ischemia and Reperfusion”, “miRNAs” and “Genic Expression”, and “miRNA”in “Kidney Transplantation” and “Ischemia and Reperfusion”, “miRNA” in “Biopsy” and “Plasma” in “Kidney Transplantation”. In relation to term “micro RNA” was found 66531 in the PUB MED, 2 COCHRANE, 1 LILACS and 74 SCIELO, with the term “Ischemia and Reperfusion”, were found 50277 in the PUB MED, 9 COCHRANE, 21 LILACS and 22 SCIELO. The search strategy involved the following databases: MEDLINE (Pub Med), LILACS, SciELO, COCHRANE, until the date 2018, march 05, according to Table 1.
| Key Words | PUBMED | COCHRANE | LILACS | SciELO |
|---|---|---|---|---|
| “miRNAs” | 66531 | 2 | 121 | 74 |
| “Rejection Acute” | 24521 | 34 | 256 | 246 |
| “Kidney Transplantation” | 117779 | 59 | 1988 | 785 |
| “Ischaemia” and “Reperfusion” | 59277 | 9 | 21 | 22 |
| “microRNAs” | 63196 | 1 | 140 | 99 |
| “miRNAs” and “Kidney Transplantation” | 188 | 2 | 0 | 0 |
| “miRNAs” and “Ischaemia and “Reperfusion” | 442 | 0 | 0 | 0 |
| “miRNAs” and “Acute Rejection Kidney” | 36 | 1 | 0 | 0 |
| “miRNA” and “Genic Expression” | 31 | 1 | 0 | 0 |
| “miRNA”in “Kidney Transplantation” and “Ischaemia and Reperfusion” | 29 | 0 | 0 | 0 |
| “miRNA”in “Biopsy”and “Plasma” in “Kidney Transplantation” | 11 | 0 | 0 | 0 |
Table 1: Search Strategy for bibliographic references.
The summary of the relevant findings is shown in Table 2 and briefly described below.
| Author (year) | Platform | Sample (s) | micro-RNAs | Main results | Reference |
|---|---|---|---|---|---|
| Anglicheau (2009) Lorenzen (2011) | qPCR qPCR | Biopsy Urine | miR-142-5p,miR- 155, miR-223, miR-30a-3p,miR-10b, let-7c miR-210, miR-10a, miR-10b | Association of acute rejection with significant changes in expression miRNAs in renal tissue grafts Potential biomarker for acute rejection and prediction of long-term GFR | [35] [36] |
| Soltaninejad (2015) | qPCR | Biopsy Blood | miR-142-5p, miR- 142-3p,miR-155,miR-223 | Diagnostics and predictive biomarkers of acute T-cell mediated rejection | [37] |
| Tao (2015) | qPCR | Serum | miR-99 a, miR-100 | Biomarkers of AR of renal grafts | [38] |
| Sui (2008) | Microarray qPCR | Biopsy | miR-320, miR-324 | Diagnostic biomarkers and probably involved in the pathogenesis of acute rejection | [39] |
| Lucille (2016) Jin (2017) | qPCR qPCR | Biopsy Urine Biopsy Serum | miR-146a miR-650 | Potential biomarker for human renal ischemia-reperfusion injury Diagnostic for acute renal rejection |
[40] [41] |
| Bijkerk (2017) | qPCR | Biopsy Serum | miR-135a, miR-199a-3p,miR-15a,miR-130b,miR-122, miR-192 | AR by systemic microvascular injury with circulating miRNA levels | [42] |
| Matz (2016) | qPCR | Biopsy Plasma | miR-15B, miR-103A, miR-106A | Discriminated stable graft function from T-cell mediated rejection | [43] |
Table 2: Main studies that tested microRNAs as diagnostic tools in clinical renal transplantation.
Lorenzen et al., evaluated the expression of miRNAs in the urinary sediment of kidney transplant recipients and found that miR-10a, miR-10b and miR-210 were differentially expressed in patients with AR in comparison to patients with stable graft function. miR-210 was found to be decreased in renal transplant patients with AR and the analysis of its expression could predict the decline of glomerular filtration rate at one year after transplantation (Lorenzen JM et al., 2011). These authors suggested that miR-210 is a potential biomarker for acute renal rejection and long-term graft function (Lorenzen JM et al., 2011).
Anglicheau et al., investigated the expression of miRNAs in the graft tissue and peripheral blood of kidney transplant recipients with AR. The authors also investigated whether these expressions would accurately predict AR of the renal graft. They found that certain miRNAs (miR-142-5p, miR-155 and miR-223) are overexpressed in renal biopsies of patients with AR and that these miRNAs are also present at higher levels in normal mononuclear cells obtained from peripheral blood. Other miRNAs were under expressed (miR-30a-3p, miR-10b or let-7c) in rejection. The authors reported that, among the 53 miRNAs differentially expressed in AR biopsies, 43 were under expressed and only 10 were overexpressed (Anglicheau D et al., 2009).
Soltaninejad et al., studied the expression levels of miR-142-5p, miR-142-3p, miR-155 and miR-223 in paired biopsy and peripheral blood mononuclear cell (PBMC) samples of renal allograft recipients with acute T-cell mediated rejection in comparison with normal allografts. The study revealed that evaluation of miR-142-3p and miR-223 expression levels in PBMC samples could be used as non-invasive predictive biomarkers of rejection and that miR-142-5p, miR-142-3p, miR-155 and miR-223 were also differentially expressed in biopsies samples of patients with rejection. Moreover, the authors suggest that a combination of a miRNAs profile should be preferred as opposed to the analysis of a single miRNA as a diagnostic tool to identifyAR (Soltaninejad E et al., 2015).
Tao et al., analyzed the miRNAs profile in serum samples from kidney transplant recipients aiming at the non-invasive detection of AR by using a miRNA array technique that was followed by quantitative real-time PCR validation. Two miRNAs, miR-99a and miR-100, were found to be up-regulated in the sera of patients with AR (Tao J et al., 2015). Interestingly, miR-99a also exhibited a potential in discriminating patients with AR from those with DGF (Tao J et al., 2015).
Sui et al., compared miRNA expression in renal biopsies of patients with acute rejection after renal transplantation with biopsies of kidney transplant recipients without rejection (Sui W et al., 2008). The authors reported an acute rejection signature of 20 miRNAs detected in 3 biopsies from patients with acute rejection as compared to 3 control renal biopsies. They found that 12 miRNAs were down-regulated and 8 miRNAs were up-regulated in the rejection samples. Furthermore, two miRNAs were validated by qRT-PCR, namely miR-320 and miR-324-3p, and were suggested to be potentially useful diagnostic biomarkers of acute rejection with perhaps a role on the pathogenesis of this process (Sui W et al., 2008).
Lucille et al., performing in vitro and in vivo experiments, identified miR-146a as a mediator of the renal response to injury that limits the consequences of inflammation (Lucile A et al., 2016). In vitro experiments in fibroblasts have shown that miR-146a is induced by senescence and acts as a negative regulator of excessive IL-6 and CXCL8 secretion, which is a hallmark of senescence (Lucile A et al., 2016). In vivo, miR-146a contributes to response after renal IRI by influencing the tubular epithelia response to inflammation via NF-kB pathway down regulation, of CXCL8 secretion, limiting post ischemic renalfibrosis (Lucile A et al., 2016).
The up regulation of miR-146a in the kidney tissue and urine of kidney transplant recipients suggests a role of miR-146a in human renal IRI (Lucile A et al., 2016). Collectively, these results emphasize miR-146a as an important effector in the pathogenesis of the renal response to IRI (Lucile A et al., 2016).
Jin et al., studied the expression and role of miR-650 in acute renal rejection in biopsies and serum in transplantation patients with acute rejection and patients with continuous stable kidney function. In patients with acute rejection was increased expression of miR-650 compared with normal allografts, suggesting with miR-650 may serve a functional role in the development of acute rejection. This biomarker regulates the B cell lymphoma -2/Bax signaling b pathway by targeting tumor suppressor inhibitor of growth protein-4, the up regulation the miR-650 contributes to the progression of acute renal rejection, occurs via promoting apoptosis and immune responses; the inhibition this biomarker provide an efficient protective effect by reducing the expression of BCL11B (transcriptional factor expressed in different types of cancer), resulting in a lower level of apoptosis and immune responses, and increased cell migration, indicating with miR-650 how potential diagnostic factor for acute renal rejection and may provide novel therapeutic strategies for its treatment (Metzinger-Le Meuth V et al., 2017).
Bijkerk et al., investigated the short- and long-term effects of AR after kidney transplantation on systemic vascular injury and the associated circulating miRNA profile. Systemic vascular injury was determined by measuring capillary tortuosity and density within the oral mucosa as by assessing circulating levels of angiopoietin-2/1 and soluble thrombomodulin.
They found an AR-associated increase in markers of systemic vascular injury, of which vascular endothelial growth factor and soluble thrombomodulin normalized within 1 year after AR. The miR-135a, miR-199a-3p and miR-15a decreased in AR and already miR-17, miR-140-3p, miR-130b, miR-122 and miR-192 increased. Of these, miR-130b, miR-199a, and miR192 associated with markers of vascular injury, whereas miR-140-3p, miR-130b, miR-122, and miR-192 normalized within 1 year after AR (BijkerkR et al., 2017).
Matz et al., evaluated the potential diagnostic value of circulating free miRNA in plasma and blood cells for rejection processes after kidney transplantation is largely unknown, but offers the potential for better and timely diagnosis of acute rejection. Free micro RNA expression of specific blood cell markers was measured in plasma from kidney transplantation with stable graft function, applying RT-PCR. The expression levels of specific microRNAs miR-15B, miR-103A and miR-106A discriminated from patients with T-cell mediated rejection (TCMR) and from patients with urinary tract infection. The measurement of circulating microRNAs in plasma from patients with renal transplants distinguishes TCMR and urinary tract infection from stable graft function. In contrast to miRNA expression measurement in blood cells it does not allow discrimination from ABMR or interstitial fibrosis and tubular atrophy (Matz M et al., 2016).
Conclusion
The discovery of miRNA has produced a giant step forward in the molecular biology field. The stability of miRNAs make them excellent candidates for non-invasive monitoring of the kidney graft in the urine and/or in the peripheral blood. There is an accumulating knowledge about diseases-specific miRNA signatures of distinct kidney graft injuries. Identification of biomarkers is crucial for improving organ allograft outcomes, as this may allow accurate prediction of an individual’s risk of graft dysfunction and individualization of immunotherapy (Godwin JG et al., 2010). Various genome-based tools are under development to predict early graft dysfunction, disease recurrence and personalized treatment options (Fleissner F, Goerzig Y, Haverich A, Thum T, 2012; Bingaman AW and Farber DL, 2004; Lorenzen JM, Haller H, Thum T, 2011 ; Mas VR et al. 2013; Van Aelst LN et al., 2016; Wang E et al., 2013).
It is expected that in the future miRNA biomarkers might became relevant, as injury markers in organ transplantation, particularly as non-invasive biomarkers of injury such as AR and IRI in kidney graft recipients. It is expected that in the future miRNA biomarkers might became relevant, as injury markers in organ transplantation, particularly as non-invasive biomarkers of injury such as AR and IRI in kidney graft recipients. Although there are so many microRNAs being studied, it is worth remembering the importance of selecting them for the study, considering important characteristics such as stability, presence in biological fluids, technique used and validation of the same, thus contributing to the research and new discoveries of targets for study.
About the Authors
Corresponding Author
Patricia Milhoransa
Renal Transplant Unit, Division of Nephrology, Hospital de ClÃnicas de Porto Alegre, Porto Alegre/RS, Brazil
- Email:
- milhoransa@ig.com.br
References
- Anglicheau D, Sharma VK, Ding R, Hummel A, et al. (2009) MicroRNA expression profiles predictive of human renal allograft status. ProcNatlAcad Sci 106: 5330-5335. https://doi.org/10.1073/pnas.0813121106
- Alarrayed S, El-Agroudy A, Al-Arrayed A, Ghareeb S, et al. (2011) Why does kidney allograft fail? A long-term single-center experience. Saudi J Kidney Dis Transpl 22: 818-824.
- Siedlecki A, Irish W, Brennan DC (2011) Delayed graft function in the kidney transplant. Am J Transplant 11: 2279-2296. https://doi.org/10.1111/j.1600-6143.2011.03754.x
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297. https://doi.org/10.1016/s0092-8674(04)00045-5
- Bustin SA, Benes V, Nolan T, PfafflMW (2005) Quantitative real-time RT-PCR- a perspective. J Mol Endocrinol 34: 597-601. https://doi.org/10.1677/jme.1.01755
- Bartel DP ( 2009 ) MicroRNAs: target recognition and regulatory functions. Cell 136: 215-233. https://doi.org/10.1016/j.cell.2009.01.002
- Balcells I, Cirera S, Busk PK (2011) Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol 11: 70. https://doi.org/10.1186/1472-6750-11-70
- Bhatt K, Mi QS, Dong Z (2011) microRNAs in kidneys: biogenesis, regulation, andpathophysiological roles. Am J Physiol Renal Physiol 300: F602-F610. https://doi.org/10.1152/ajprenal.00727.2010
- Bijkerk R, van Solingen C, de Boer HC, de Vries DK, et al. (2014) Silencing of miRNA-126 in kidney ischemia reperfusion is associated with elevated SDF-1 levels and mobilization of Sca-1+/Lin- progenitor cells. Microrna 3: 144-149. https://doi.org/10.2174/2211536604666150121000340
- Chang TC and Mendell JT (2007) microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet 8: 215-239. https://doi.org/10.1146/annurev.genom.8.080706.092351
- Aquino-Dias EC, Joelsons G, da Silva DM, Berdichevski RH, et al. (2008) Non-invasive diagnosis of acute rejection in kidney transplants with delayed graft function. Kidney Int 73: 877–884. https://doi.org/10.1038/sj.ki.5002795
- Eltzschig HK and Eckle T (2011) Ischemia and reperfusion – from mechanism totranslation. Nat Med 17: 1391-1401. https://doi.org/10.1038/nm.2507
- Soltaninejad E, Nicknam MH, Nafar M, Ahmadpoor P, et al. (2015) Differential expression of microRNAs in renal transplant patients with acute T-cell mediated rejection. TransplImmunol 33: 1-6. https://doi.org/10.1016/j.trim.2015.05.002
- Fleissner F, Goerzig Y, Haverich A, Thum T (2012) Microvesicles as novel biomarkers and therapeutic targets in transplantation medicine. Am J Transplant 12: 289-297. https://doi.org/10.1111/j.1600-6143.2011.03790.x
- Geddes CC, Woo YM, Jardine AG (2002) The impact of delayed graft function on the long-term outcome of renal transplantation. J Nephrol 15: 17-21.
- Girlanda R and Kirk AD (2007) Frontiers in nephrology: immune tolerance to allografts in humans. J Am SocNephrol 18: 2242-2251. https://doi.org/10.1681/asn.2007020180
- GwinnerW (2007) Renal transplant rejection markers. World J Urol 5: 445-455. https://doi.org/10.1007/s00345-007-0211-6
- Godwin JG, Ge X, Stephan K, Jurisch A, et al. (2010) Identification of a microRNA signature of renal ischemia reperfusion injury. ProcNatlAcadSci 107: 14339-14344. https://doi.org/10.1073/pnas.0912701107
- Harris A, Krams SM, Martinez OM (2010) MicroRNAs as immune regulators: implications for transplantation. Am J Transplant. 10: 713-719. https://doi.org/10.1111/j.1600-6143.2010.03032.x
- Hartono C, Muthukumar T, Suthanthiran M (2010) Noninvasive diagnosis of acuterejection of renal allografts. CurrOpin Organ Transplant 15: 35-41. https://doi.org/10.1097/mot.0b013e3283342728
- Huang X, Le QT, Giaccia AJ (2010 ) Mir-210-micromanager of the hypoxia pathway. Trends Mol Med 16: 230-237. https://doi.org/10.1016/j.molmed.2010.03.004
- Bingaman AW and Farber DL (2004) Memory T cells in transplantation: generation, function, and potential role in rejection. Am J Transplant 4: 846–852. https://doi.org/10.1111/j.1600-6143.2004.00453.x
- Kim DH, Saetrom P, Snøve O Jr, Rossi JJ (2008) MicroRNA-directed transcriptionalgene silencing in mammalian cells. Proc Natl Acad Sci 105: 16230-16235. https://doi.org/10.1073/pnas.0808830105
- Solez K, Colvin RB, Racusen LC, Haas M, et al. (2008) Banff 07 classification of renalallograft pathology: updates and future directions. Am J Transplant 8: 753-760. https://doi.org/10.1111/j.1600-6143.2008.02159.x
- Kato M, Putta S, Wang M, Yuan H, et al. (2009) TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 11: 881-889. https://doi.org/10.1038/ncb1897
- Lee SO, Masyuk T, Splinter P, Banales JM, et al. (2008) MicroRNA 15a modulates expression of the cell-cycle regulator Cdc25A and affectshepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest. 118: 3714-3724. https://doi.org/10.1172/jci34922
- Li JY, Yong TY, Michael MZ, Gleadle JM (2010) Review: The role of microRNAs in kidney disease. Nephrology (Carlton) 15: 599-608. https://doi.org/10.1111/j.1440-1797.2010.01363.x
- Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods 25: 402-408. https://doi.org/10.1006/meth.2001.1262
- Lorenzen JM, Haller H, Thum T (2011) MicroRNAs as mediators and therapeutic targets in chronic kidney disease. Nat Rev Nephrol. 7: 286-294. https://doi.org/10.1038/nrneph.2011.26
- Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, et al. (2011) Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 11: 2221-2227. https://doi.org/10.1111/j.1600-6143.2011.03679.x
- Lorenzen JM (2015) Vascular and circulating microRNAs in renal ischaemia-reperfusion injury. J Physiol 593: 1777-17784. https://doi.org/10.1113/jp270318
- Lucile A, Geoffroy D, Marion R, Sauvaget V et al., (2016) MicroRNA-146a in Human and Experimental Ischemic AKI: CXCL8-Dependent Mechanism of Action. J Am SocNephrol 28: 479-493. https://doi.org/10.1681/asn.2016010045
- Meier-Kriesche HU, Ojo AO, Port FK, Arndorfer JA, et al. (2001) Survival improvement among patients with end-stage renal disease: trends overtime for transplant recipients and wait-listed patients. J Am SocNephrol 12 : 1293-1296.
- Melkonyan HS, Feaver WJ, Meyer E, Scheinker V, et al. (2008) Transrenal nucleic acids: from proof of principle to clinical tests. Ann N Y Acad Sci 1137: 73-81. https://doi.org/10.1196/annals.1448.015
- Mas VR, Dumur CI, Scian MJ, Gehrau RC, et al. (2013) MicroRNAs as biomarkers insolid organ transplantation. Am J Transplant 13:11-19. https://doi.org/10.1111/j.1600-6143.2012.04313.x
- Marí-Alexandre J, Sánchez-Izquierdo D, Gilabert-Estellés J, Barceló-Molina M, et al. (2016) miRNAs Regulation and Its Role as Biomarkers inEndometriosis. Int J Mol Sci 17: 93. https://doi.org/10.3390/ijms17010093
- Newell KA, Asare A, Kirk AD, Gisler TD, et al. (2010) Immune Tolerance Network ST507 Study Group. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest 6: 1836-1847. https://doi.org/10.1172/jci39933
- O’Connell RM, Tagano KD, Boldin MP, Cheng G, et al. (2007) MicroRNA-155 is induced during the macrophage inflammatory response. ProcNatlAcadSci 104:1604-1609. https://doi.org/10.1073/pnas.0610731104
- Grosso G, Corona D, Mistretta A, Zerbo D, et al. (2012) Delayed graft function and long-term outcome in kidney transplantation. Transplant Proc 44: 1879–1883. https://doi.org/10.1016/j.transproceed.2012.06.044
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901-906. https://doi.org/10.1038/35002607
- Serón D, Alexopoulos E, Raftery MJ, Hartley RB, et al. (1989) Diagnosis ofrejection in renal allograft biopsies using the presence of activated andproliferating cells. Transplantation 5: 811-816. https://doi.org/10.1097/00007890-198905000-00013
- Suthanthiran M (1998) Human renal allograft rejection: molecular characterization. Nephrol DialTransplant 13: 21– 24. https://doi.org/10.1093/ndt/13.suppl_1.21
- Haas M, Sis B, Racusen LC, Solez K, et al. (2014) Banff meeting report writing committee. Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. Am J Transplant 14: 272-283. https://doi.org/10.1111/ajt.12590
- Sui W, Dai Y, Huang Y, Lan H, et al. (2008) Microarray analysis of MicroRNA expression inacute rejection after renal transplantation. Transpl Immunol 19: 81–85. https://doi.org/10.1016/j.trim.2008.01.007
- Shan J, Feng L, Luo L, Wu W, et al. (2011) MicroRNAs: Potential biomarker in organ transplantation. Transplant Immunol 24: 210-215. https://doi.org/10.1016/j.trim.2011.03.004
- Spiegel JC, Lorenzen JM, Thum T (2011) Role of microRNAs in immunity and organ transplantation. Expert ReviMol Medicine 13:e37. https://doi.org/10.1017/s1462399411002080
- Sarma NJ, Tiriveedhi V, Ramachandran S, Crippin J, et al. (2012) Modulation of immune responses following solid organ transplantation by microRNA. ExpMolPathol 3: 378-385. https://doi.org/10.1016/j.yexmp.2012.09.020
- Tao J, Yang X, Han Z, Lu P, et al. (2015) Serum MicroRNA-99a Helps Detect Acute Rejection in Renal Transplantation. Transplantat Proceed 47:1683-1687. https://doi.org/10.1016/j.transproceed.2015.04.094
- Van Aelst LN, Summer G, Li S, Gupta SK, et al. (2016) RNA Profiling in human and murine transplanted hearts: identification and validation of therapeutic targets for acute cardiac and renal allograft rejection. Am J Transpl 16: 99-110. https://doi.org/10.1111/ajt.13421
- Wang E, Nie Y, Zhao Q, Wang W, et al. (2013) Circulating miRNAs reflect early myocardial injury and recovery after heart transplantation. J Cardiothorac Surgery 8: 1-26. https://doi.org/10.1186/1749-8090-8-165
- Nassirpour R, Raj D, Townsend R, Argyropoulos C (2016) MicroRNA biomarkers in clinical renal disease: from diabete nephropathy renal transplantation and beyond. Food ChemToxicol 98: 73-88. https://doi.org/10.1016/j.fct.2016.02.018
- Metzinger-Le Meuth V, Burtey S, Maitrias P, Massy ZA, et al. (2017) microRNAsin the pathophysiology of CKD-MBD: Biomarkers and innovative drugs. Biochim Biophys Acta Mol Basis Dis 1863: 337-345. https://doi.org/10.1016/j.bbadis.2016.10.027
- Ulbing M, Kirsch AH, Leber B, Lemesch S, et al. (2017) MicroRNAs 223-3p and 93-5p in patients with chronic kidney disease before and after renal transplantation. Bone 95: 115-123. https://doi.org/10.1016/j.bone.2016.11.016
- Bijkerk R, Florijn BW, Khairoun M, Duijs JMGJ, et al. (2017) Acute Rejection After Kidney Transplantation Associates With Circulating MicroRNAs and Vascular Injury. Transplant Direct 3: e174. https://doi.org/10.1097/txd.0000000000000699
- Matz M, Lorkowski C, Fabritius K, Durek P, et al. (2016) Free microRNA levels in plasma distinguish T-cell mediated jection from stable graft function after kidney transplantation. TransplImmunol 39: 52-59. https://doi.org/10.1016/j.trim.2016.09.001
- Abbas A K, Lichtman A H, Pillai S (2006) Imunolgia Celular e Molecular. 5 edn, Rio de Janeiro, Elsevier, 580 p.
Keywords:
Download:
Full PDF- Share This