Marker-Assisted elimination of drought-susceptible accessions in upland rice breeding
Received: January 14, 2018
Accepted: February 24, 2018
Published: February 27, 2018
Genet.Mol.Res. 17(1): gmr16039886
DOI: 10.4238/gmr16039886
Abstract
Breeding for water-deficit tolerance is fundamental to guarantee the sustainability of upland rice production, mainly due to the possibility of an increased frequency of drought episodes due to climate change. This work aimed to identify single nucleotide polymorphism (SNP) markers, derived from RNA sequencing (RNA-Seq), genome-wide association study (GWAS) and candidate genes from Arabidopsis, with potential for use in marker-assisted selection (MAS) for drought tolerance. RNA-Seq and GWAS were efficient in identifying useful SNP markers from the data obtained from three years of field experiments for 175 upland rice accessions, which were sequenced using 32 genes by Capture-Seq. Three genes were equally able to generate SNP markers that discriminated 95% of the 20 most drought susceptible accessions in the joint analysis of the experiments. The elimination of the genotypes with the unfavourable SNP allele reduced the initial number of accessions to one third, and transferring this result in a breeding routine, would enable to conduct smaller experiments per target location, increasing the precision and reducing the cost of the drought phenotyping.
Introduction
Drought is one of the major problems affecting crop yields, particularly for upland rice (Oryza sativa L.), mainly when cultivated in areas that depend exclusively on rainfall, resulting in low average grain yield (Serraj et al., 2011). Drought in rainy areas is expected to increase due to climate change, which can cause losses in productivity, especially if it occurs in the reproductive stage (Fedoroff et al., 2010; Kumar et al., 2014). In Brazil, the upland rice cultivated area has decreased by 57% in the last decade due to area competition with corn and soybean, and due to the lower yields (1.8 ton/ha) compared to lowland rice (7.0 ton/ha), contributing only about 38% of Brazilian rice production (Barbosa Filho and Yamada 2002; Heinemann et al., 2015). However, rice production in the lowlands can decrease due to reduced water availability around the world and, in this scenario, upland rice is a great alternative. To be feasible, future upland rice cultivars should combine the characteristics of resistance to drought in couple with high productivity when cultivated under conditions of water availability (Serraj et al., 2009; Fageria et al., 2014).
In upland rice, genetic variation for drought tolerance is available in gene banks (Pantalião et al., 2016), and the useful variability must be better exploited in order to be incorporated into the breeding programs. (Serraj et al., 2011; Kumar et al., 2014). However, the development of drought tolerant cultivars, combining high productive potential and drought tolerance is difficult, since drought is a quantitative trait, with low heritability, environmental interaction, and confounding effect with other abiotic stresses (Swamy and Kumar, 2013). Despite these issues, a great deal of effort has been carried out to obtain drought tolerant lines and cultivars, focusing in experiments in target environments, and the selection of many genotypes throughout the seasons and years (Degenkolbe et al., 2013; Swamy and Kumar, 2013).
Much information on the structural and functional genome of rice is publicly available, and such information can be used as a starting point for studies on various traits of interest. Genetic mapping, physiological characterization and expression analyses are available to potentially identify genes that have major effects on grain yield under drought treatment (Swamy et al., 2013). In addition to the development of drought tolerant cultivars by breeding programs, an important strategy would be to use recent scientific advances in biotechnology to improve this trait (Serraj et al., 2011). The complex genetic nature of drought tolerance, which hampers classic breeding, and poor understanding of the physiological and molecular mechanisms associated with this trait, make also the molecular breeding a difficult task.
Genes related to drought tolerance in rice have recently been obtained via mapping of QTLs (Dixit et al., 2014) and association mapping (Pantalião et al., 2016) by means of single nucleotide polymorphism (SNP) markers, functional genomics, microarrays (Parida et al., 2012; Borah et al., 2017), RNA-Seq (Huang et al., 2014; Silveira et al., 2015) or simply by using candidate genes homologous to those identified in other species (Shaar-Moshe et al., 2015; Abreu et al., 2016). The cost of high-resolution genotyping, as well as sequencing, has fallen year by year, which has opened the possibility of identifying genes related to drought tolerance via whole-genome resequencing (Jain et al., 2014). Nevertheless, the selection of drought-tolerant material based on these markers is still not a routine practice in marker-assisted selection (MAS) because of their inconsistency due to large QTL × E interaction, and no clear evidence exists that these markers have been successful in improving yield under drought stress in rice (Bernier et al., 2009; Swamy and Kumar, 2013). Most efforts to improve the drought tolerance of rice through MAS have been devoted to the use of low-resolution QTLs represented by large chromosomal segments, which only capture a small proportion of the total phenotypic variation (Babu, 2010). The possible reasons for this could be the inconsistency of QTL effects when considering multiple environments, and the detection of small effect QTLs on grain yield (Serraj et al., 2011; Dixit et al., 2014).
In the present work, a set of 175 upland rice genotypes (tropical japonica) were evaluated in three years of field experiments under water deficit, and also sequenced for a series of genes previously identified as being related to drought response. This work aimed to develop a strategy to select SNPs useful for a marker-assisted selection for drought tolerance in rice.
Material and Methods
Plant material
A set of 175 upland rice (Oryza sativa ssp. japonica) accessions from the Embrapa Rice Core Collection (Abadie et al., 2005) was evaluated, being 83 Brazilian landraces (acronym LRC), 47 improved varieties from Brazil (IVB) and 45 improved varieties from other countries (IVC), in addition to three checks (BRS Esmeralda, BRS Soberana and BRSGO Serra Dourada) (Supplemental Table 1).
| Gene | ID RGAP | Size (bp) | SNPs* |
|---|---|---|---|
| 1 | LOC_Os01g28500 | 766 | 12 |
| 2 | LOC_Os01g57854 | 6655 | 2 |
| 3 | LOC_Os03g08320 | 1551 | 11 |
| 4 | LOC_Os04g01740 | 2825 | 4 |
| 5 | LOC_Os04g39420 | 2377 | 1 |
| 6 | LOC_Os04g40990 | 2491 | 28 |
| 7 | LOC_Os05g50380 | 5484 | 1 |
| 8 | LOC_Os09g17560 | 1572 | 8 |
| 9 | LOC_Os09g21120 | 2592 | 11 (10) |
| 10 | LOC_Os09g21230 | 4532 | 2 |
| 11 | LOC_Os09g31486 | 3215 | 1 |
| 12 | LOC_Os10g31780 | 786 | 35 |
| Total SNPs from RNA-Seq | 116 | ||
| 13 | LOC_Os01g43060 | 2535 | 2 |
| 14 | LOC_Os02g09650 | 1313 | 30 (6) |
| 15 | LOC_Os02g43330 | 1423 | 5 |
| 16 | LOC_Os03g50570 | 5281 | 1 |
| 17 | LOC_Os04g33710 | 747 | 10 |
| 18 | LOC_Os06g40570 | 4903 | 1 |
| 19 | LOC_Os06g41580 | 435 | 33 |
| 20 | LOC_Os06g41690 | 11321 | 1 |
| 21 | LOC_Os06g41810 | 3008 | 1 |
| 22 | LOC_Os08g13320 | 4706 | 1 |
| 23 | LOC_Os08g13430 | 2633 | 5 |
| 24 | LOC_Os08g13560 | 2476 | 13 |
| 25 | LOC_Os08g13840 | 1144 | 30 (9) |
| 26 | LOC_Os11g31950 | 13853 | 1 |
| Total SNPs from GWAS | 134 | ||
| EIN | LOC_Os07g06130 | 7031 | 1 |
| GLUR | LOC_Os02g02540 | 5174 | 1 |
| CESA | LOC_Os01g54620 | 5743 | 2 |
| PLD | LOC_Os01g07760 | 4971 | 1 |
| OSKH2 | LOC_Os06g08450 | 8126 | 19 |
| LSI1 | LOC_Os02g51110 | 3716 | 2 |
| Total SNPs from candidate genes | 26 | ||
| Total | 276 | ||
* Between parenthesis: Number of SNPs able to discriminate most of the tolerant and susceptible upland rice accessions.
Table 1: List of the 32 drought-related genes identified by RNA-Seq (1 to 12), GWAS (13 to 26) and candidate genes (27 to 32), sequenced by Capture-Seq.
Field trials assessing drought stress
The experiments were conducted in three years (2010, 2011 and 2012) at the Drought Phenotyping Experimental Station, located in Porangatu, Goias State, Brazil (49o06′W, 13o18′S, 396 m of altitude). For 2010 and 2011, the experimental design was Federer augmented blocks (FABs), and for 2012, it was randomized complete blocks, with two replications. The plot size consisted of four rows of three meters with a density of 60 seeds per meter. The experiments were monitored by tensiometers at a depth of 15 cm to maintain suitable soil water conditions (- 0.025 MPa). The drought was applied 30 days after emergence, when irrigation was reduced to approximately 50% until the end of the experiment. After the complete physiological maturity of the grains, the rice plants were manually harvested. Statistical analysis of grain yield data (kg/ha) was performed using the lme4 package from R software version 3.0.1 (The R Foundation for Statistical Computing, 2015) using the mixed model analysis procedure. Coefficient of variation (CV), accuracy and heritability for yield were estimated according to Resende and Duarte (2007). Estimates of the variance components were obtained by the REML (restricted maximum likelihood) method, according to Patterson and Thompson (1971), and the genetic values of everyone were estimated using the empirical best linear unbiased predictor. The genotype stability was estimated by the harmonic mean of the genotypic values (HMGV), and the adaptability was estimated by the relative performance of the genotypic values (RPGV), whereas the harmonic mean of the relative performance of genotypic values (HMRPGV) evaluated both the adaptability and stability of the genotypes (Resende, 2004).
DNA extraction and capture sequencing probe design
The DNA of 175 upland rice genotypes was obtained from the leaf tissue of seedlings using the Biopur Mini Spin Plant Extraction Kit (www.biometrix.com.br). The DNA samples were quantified on a Nanodrop spectrophotometer (ThermoScientific) and diluted to a concentration of 40 ng/μL. The integrity of the samples was assessed on 1% agarose gel. The probe set used for Capture-Seq were designed according Neves et al. (2013) for 32 genes (Table 1) whose selection was based on studies with RNA-Seq (Silveira et al., 2015), a genome-wide association study (Pantalião et al., 2016) and candidate genes (Abreu et al., 2016).
Capture sequencing analysis
The 175 upland rice genotypes were sequenced using the Capture-Seq methodology described in Neves et al. (2014). The DNA samples were sent to Rapid Genomics (USA) for capture sequencing (Capture-Seq). For each sequenced gene, a fragment of 120 nucleotides was selected and complimentary biotinylated RNA probes where synthesized. Prior to the enrichment step, Illumina-comparable libraries where prepared for each sample (Bentley et al., 2008). An input of 500 ng of DNA was used and fragmented to an average of 300 bp. Libraries were constructed by end-repairing the sheared DNA, A-tailing, ligating standard adapters to each molecule and enriching the libraries for ligated fragments with PCR amplification. The libraries were multiplexed prior to hybridization and 750 ng of pooled libraries were combined with the probes to enrich for the genes of interest following Sure Select Target Enrichment protocol (Agilent Technologies). The library pool was sequenced on a HiSeq 3000 platform (Illumina) according to the manufacturer's recommendations, with 100 cycles with paired-end fragments. Sequencing data was converted to fastq files using Illumina’s bcl2fastq software on default settings.
Identification of drought-related SNPs
The sequences from Capture-Seq analysis were trimmed by removing the low-quality bases, using the parameter phred < 20 at the 3' ends of each sequence in the Trimmomatic tool. MosaikAligner and Freebayes were used to align and call SNPs, respectively. To identify markers related to tolerance or susceptibility to drought, grain yield data from the 2012 experiment were used, since the genotypes were evaluated with repetition. The SNP profiles of the 20 genotypes with the highest yield and 20 genotypes with the lowest yield were compared.
Results
Drought phenotyping
The coefficient of variation, accuracy and heritability for yield for the 2010 experiment was 14%, 0.85% and 0.62%, respectively; for 2011, these values were 11%, 0.77% and 0.51%; and for 2012, they were 16%, 0.86% and 0.61%. Although the experimental design of the 2010 and 2011 trials was in FABs, these values were very similar to the values found in the 2012 experiment, which used randomized blocks, enabling direct comparisons among genotype and yield performances. The F test of the joint analysis of variance showed that the effects of year and year × genotypes were significant (p<0.001) for grain yield. Accessions above the yield average for the three field experiments were found for all groups of accessions (LRC, IVB and IVC). This means that the selection of drought-tolerant materials should not necessarily be prioritized in landraces, which are known to have unwanted traits for rice commercial cultivars, such as long cycle, tall size, and lower grain quality for trade.
Capture sequencing
The 175 individuals sequenced for the 32 genes generated a total of 276 SNPs. The set of genes with the highest number of SNPs identified were derived from GWAS, with 134 SNPs (mean of 9.6 SNPs per gene), the same average obtained by the set of SNPs from RNA-Seq. The set derived from candidate genes showed only 26 SNPs (19 of these SNPs came from LOC_Os06g08450) and the lowest average of SNP number per gene (4.3) (Table 1). The gene with the highest number of SNPs identified was LOC_Os10g31780 (35 SNPs, from RNA-Seq), whereas the gene with the highest SNP density was LOC_Os06g40570 (13 bp/SNP, from GWAS).
Drought-related SNPs
Considering all 276 SNPs found for 32 genes, 15 SNPs were informative (which were discriminant for the 20 drought-tolerant and 20 drought-susceptible accessions) from GWAS series (9 from LOC_Os08g13840 and 6 from LOC_Os02g09650) and 10 SNPs were informative from RNA-Seq series (all from LOC_Os09g21120) (Table 1). These three genes were located at positions S2_4959308, S8_8258609 and S9_12748551, respectively (from Nippon bare reference genome, available at RGAP website - http://rice.plantbiology.msu.edu). These SNPs presented the same classification pattern for the 20 most tolerant and susceptible genotypes (Table 2), i.e., they showed the AA SNP pattern for the same tolerant accessions, and for that reason, a single SNP (S2_4959308, S2 acronym, here-after) was considered from now on.
| Accession | 2012 yield (Kg/ha) | S2_4959308 | S8_8258609 | S9_12748551 | |
|---|---|---|---|---|---|
| 20+ | |||||
| 1 | CascaBranca | 3186.46 | AA | AA | AA |
| 2 | Aimoré | 3071.74 | AA | AA | AA |
| 3 | Guarani | 2924.01 | AA | AA | AA |
| 4 | Tox 514-16-101-1 | 2916.93 | GG | TT | CC |
| 5 | Carajás | 2903.85 | AA | AA | AA |
| 6 | Uruçuí | 2843.56 | AA | AA | AA |
| 7 | ArrozCarolino | 2797.83 | AA | AA | AA |
| 8 | Ct11891-3-3-3-M | 2651.36 | AA | AA | AA |
| 9 | Irat 112 | 2616.63 | AA | AA | AA |
| 10 | Tangará | 2492.42 | AA | AA | AA |
| 11 | Arroz 4 Meses | 2381.64 | AA | AA | AA |
| 12 | TrêsMesesBranco | 2286.65 | AA | AA | AA |
| 13 | Guaíra | 2226.53 | AA | AA | AA |
| 14 | Branquinho 90 Dias | 2218.80 | AA | AA | AA |
| 15 | ArrozBrancoPrecoce | 2217.43 | GG | TT | CC |
| 16 | A12-286-1-1 | 2198.67 | GG | TT | CC |
| 17 | Arroz 3 Meses | 2169.66 | GG | TT | CC |
| 18 | Brs Primavera | 2164.04 | GG | TT | CC |
| 19 | CatetoAmarelo | 2114.27 | GG | TT | CC |
| 20 | 1r65907-188-1-B | 2104.40 | GG | TT | CC |
| 20- | |||||
| 1 | Arroz Da Terra | 626.63 | AA | AA | AA |
| 2 | Cana Roxa | 625.37 | AA | AA | AA |
| 3 | TrêsPotes | 619.10 | GG | TT | CC |
| 4 | Ipeaco11p | 617.44 | GG | TT | CC |
| 5 | ArrozMatoGrosso | 615.70 | GG | TT | CC |
| 6 | N.7384 [Rpl× Daniela] | 610.65 | GG | TT | CC |
| 7 | Br4742-B-19-23 | 577.94 | GG | TT | CC |
| 8 | Ct10037-9-4-M-1-1p-2-M | 554.41 | AA | AA | AA |
| 9 | Yunlu N 1 | 548.24 | GG | TT | CC |
| 10 | BicoPreto | 546.96 | AA | AA | AA |
| 11 | Tb47h-MR-11-51-3 | 539.85 | GG | TT | CC |
| 12 | Ls 85-158 | 534.47 | GG | TT | CC |
| 13 | BaePelado | 529.01 | AA | AA | AA |
| 14 | Arroz De Guerra | 515.38 | GG | TT | CC |
| 15 | Vermelhão | 506.35 | GG | TT | CC |
| 16 | Cabaçu | 492.63 | GG | TT | CC |
| 17 | Honduras | 442.74 | GG | TT | CC |
| 18 | Yn1906-Uul 65 | 435.39 | GG | TT | CC |
| 19 | Blue Belle | 413.83 | GG | TT | CC |
| 20 | Farroupilha | 358.39 | AA | AA | AA |
Table 2: SNP profile of the 20 most productive (20+) and 20 least productive (20-) accessions (2012 drought experiment in the field) for the three selected markers.
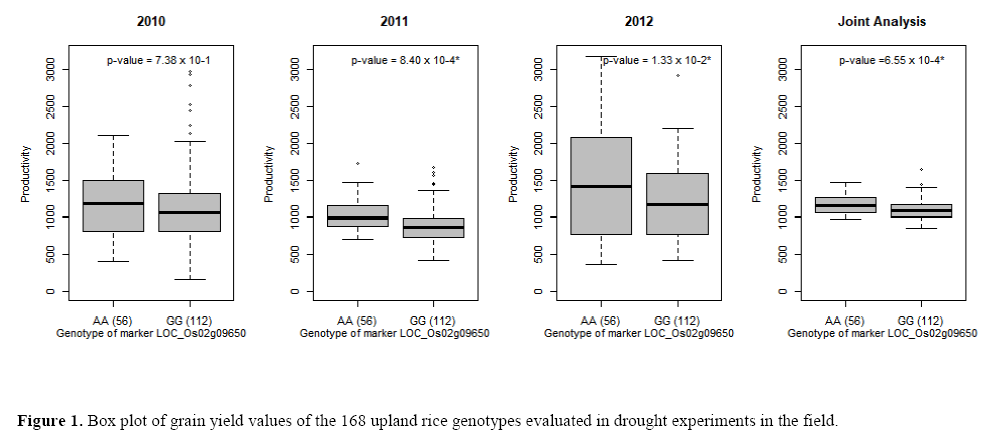
Of the 175 accessions evaluated with the SNP S2, 60 were genotyped as AA and 115 as GG. In seven accessions, was not possible to obtain the grain yield data for all three years, and for this reason, they were removed from the yield joint analysis. Considering the 168 remaining accessions, in all experiments and joint analysis the average yield of 56 AA genotypes were significantly higher than the average for 112 GG genotypes (Figure 1).
The percentage of SNP S2 in selecting genotypes with an above-average yield for two or three years (selection for AA genotypes) was just 50%. On the other hand, considering the genotypes with below-average yield for two or three years (selection for GG genotypes), the percentage was 76 and 80%, respectively (Table 3).
| Variables | Occurrences | SNPS2 success |
|---|---|---|
| Accessions with yield above the average for 3 years | 20 | 50% |
| Accessions with yield above the average for 2 years | 46 | 50% |
| Accessions with yield below the average for 3 years | 35 | 80% |
| Accessions with yield below the average for 2 years | 67 | 76% |
| Total | 168 |
Table 3: Success of SNP S2 (AA or GG patterns) to predict the yield of accessions above or below the average in three years of drought experiments
The setup of three drought tolerance experiments showed to be fundamental to discriminate accessions with variable performance for this trait. For example, the cultivars Primavera and Soberana are known to be susceptible to drought, but in one experiment, their yield was above average. When the phenotypic and genotypic results were compared in the pairwise analysis of the experiments, the percentage of SNP S2 to discriminate for drought tolerance ranged from 45% (2011 × 2012) to 62% (2010 × 2012), whereas for susceptibility, it varied from 61% (2010 × 2012) to 87% (2010 × 2011) (Table 4).
| Variables | Number of accessions | SNP S2 Coincidence | Distribution | SNP S2 Coincidence |
|---|---|---|---|---|
| 2010=2011=2012 | 55 | 69% | 20 A | 50% |
| 2010=2011≠2012 | 36 | 72% | 13 A | 46% |
| 2010=2012≠2011 | 36 | 61% | 13 A | 62% |
| 2011=2012≠2010 | 41 | 63% | 20 A | 45% |
| Total | 168 |
Table 4: Coincidence percentages between yield and the SNP S2 pattern (AA or GG) for 168 upland rice accessions evaluated in three years of drought experiments. A: above the yield average; B: Below the yield average.
The high percentage of susceptible accessions identified by AA allele of the SNP S2 indicates that selection to discard GG accessions from the total set of accessions is advantageous for breeding purposes. Instead of evaluating 168 accessions, the 56 accessions with the AA genotype identified by SNP S2 would be selected, that is, only 33.3% of the accessions, and these would then be evaluated in smaller experiments, with a higher number of repetitions in the environment for which the drought-tolerant cultivars under selection are intended. Of these 56 AA selected accessions, 38 (68%) are landraces (LRC), while 9 (16%) are improved varieties from Brazil (IVB) and 9 (16%) are improved varieties from other countries (IVC). For the 112 GG accessions, the accessions were better distributed among groups, with 42 (38%) LRC, 36 (32%) are IVC and 34 (30%) are IVB.
Adaptability and stability analysis for drought yield
The parameter used to determine genotype adaptability was RPGV; HMGV was used to determine stability, whereas HMRPGV evaluated both adaptability and stability. The classification of genotypes was based on adaptability since the aim is to identify a genotype that is locally adapted to drought. However, the results were quite similar between the different parameters, including the yield (empirical best linear unbiased predictor, Eblups) of the joint analysis (Table 5). Considering the 20 most adapted genotypes, 8 are LRC, 7 are IVB and 5 are IVC; among the 20 least adapted, 9 are LRC, 5 IVB and 6 IVC (Table 5). SNP S2 genotyping analysis showed that 19 of the 20 drought susceptible genotypes (or 95%) in the joint analysis have the GG pattern.
| Genotype | Group | SNP S2 | RPGV | RPGV*GA | HMGV | HMRPGV | HMRPGV*GA | BLUPj |
|---|---|---|---|---|---|---|---|---|
| 20+ | ||||||||
| IRAT 141 | IVC | GG | 2.20 | 2426.44 (1) | 2069.38 (1) | 1.95 | 2160.27 (1) | 1647.72 (1) |
| CascaBranca | LRC | AA | 1.77 | 1954.78 (2) | 1779.14 (2) | 1.71 | 1884.11 (2) | 1474.76 (2) |
| Aimoré | IVB | AA | 1.74 | 1928.03 (3) | 1752.17 (4) | 1.66 | 1839.51 (4) | 1458.90 (3) |
| TOX 514 | IVC | GG | 1.71 | 1886.57 (4) | 1728.04 (5) | 1.66 | 1838.14 (5) | 1446.17 (4) |
| IRAT 112 | IVC | AA | 1.70 | 1879.77 (5) | 1762.01 (3) | 1.68 | 1860.22 (3) | 1435.91 (5) |
| Uruçuí | IVB | AA | 1.65 | 1822.95 (6) | 1668.32 (7) | 1.60 | 1765.39 (6) | 1418.15 (6) |
| Jaguary | LRC | GG | 1.64 | 1817.51 (7) | 1370.41 (22) | 1.24 | 1365.49 (29) | 1382.09 (11) |
| BGA 5975 | IVB | GG | 1.64 | 1808.20 (8) | 1531.93 (16) | 1.47 | 1627.42 (13) | 1402.26 (8) |
| CT11891 | IVC | AA | 1.63 | 1802.20 (9) | 1685.73 (6) | 1.60 | 1762.99 (7) | 1400.38 (9) |
| Carajás | IVB | AA | 1.60 | 1770.63 (10) | 1594.86 (10) | 1.52 | 1682.36 (10) | 1396.91 (10) |
| Carolino | LRC | AA | 1.59 | 1759.46 (11) | 1502.35 (17) | 1.48 | 1638.78 (12) | 1405.66 (7) |
| CT10006 | IVC | GG | 1.56 | 1719.45 (12) | 1562.05 (13) | 1.47 | 1623.51 (14) | 1355.89 (14) |
| 3 Meses | LRC | GG | 1.55 | 1714.20 (13) | 1645.08 (9) | 1.53 | 1694.02 (9) | 1351.00 (15) |
| IAC 165 | IVB | GG | 1.55 | 1710.19 (14) | 1658.01 (8) | 1.54 | 1706.04 (8) | 1347.21 (16) |
| Guarani | IVB | AA | 1.53 | 1686.42 (15) | 1416.83 (20) | 1.40 | 1547.43 (18) | 1378.76 (12) |
| Tangará | IVB | AA | 1.52 | 1682.74 (16) | 1544.41 (14) | 1.49 | 1643.59 (11) | 1359.99 (13) |
| Cacho Grande | LRC | AA | 1.50 | 1660.24 (17) | 1575.32 (11) | 1.45 | 1604.95 (17) | 1321.32 (17) |
| Agulha | LRC | GG | 1.49 | 1644.78 (18) | 1348.38 (27) | 1.23 | 1358.46 (31) | 1314.39 (20) |
| Noventinha | LRC | AA | 1.47 | 1623.29 (19) | 1563.59 (12) | 1.47 | 1621.09 (15) | 1317.54 (18) |
| Japonês Grande | LRC | GG | 1.46 | 1610.46 (20) | 1542.52 (15) | 1.46 | 1608.90 (16) | 1315.43 (19) |
| 20 - | ||||||||
| Vermelhinho | LRC | GG | 0.74 | 817.01 (149) | 779.18 (141) | 0.73 | 806.58 (136) | 987.69 (146) |
| Catetinho | LRC | GG | 0.74 | 814.24 (150) | 785.34 (136) | 0.72 | 793.29 (139) | 977.98 (151) |
| IREM 238 | IVC | GG | 0.73 | 809.06 (151) | 767.15 (147) | 0.70 | 773.14 (149) | 975.76 (153) |
| Amarelão | LRC | AA | 0.73 | 808.69 (152) | 615.28 (163) | 0.62 | 689.09 (158) | 988.49 (145) |
| Catetinho | LRC | GG | 0.73 | 806.24 (153) | 749.55 (151) | 0.72 | 791.80 (141) | 987.30 (147) |
| IAC 81-176 | IVB | GG | 0.73 | 801.80 (154) | 770.12 (146) | 0.71 | 780.51 (144) | 974.12 (155) |
| CNAX 1503-12-9-4-B | IVB | GG | 0.72 | 799.95 (155) | 777.99 (142) | 0.72 | 794.31 (138) | 975.32 (154) |
| Arcos Branco | LRC | GG | 0.71 | 779.82 (156) | 743.79 (152) | 0.70 | 775.29 (146) | 973.01 (156) |
| LigeiroVermelho | LRC | GG | 0.70 | 777.91 (157) | 752.62 (149) | 0.69 | 757.57 (153) | 962.20 (158) |
| LS 85-125 | IVB | GG | 0.69 | 763.37 (158) | 696.02 (158) | 0.61 | 677.73 (160) | 945.65 (161) |
| L 285 | IVC | GG | 0.68 | 754.89 (159) | 700.69 (156) | 0.67 | 741.88 (155) | 968.74 (157) |
| IRAT 142 | IVC | GG | 0.66 | 724.70 (160) | 700.68 (157) | 0.65 | 716.98 (157) | 945.61 (162) |
| Rio Paraguay | IVB | GG | 0.65 | 716.93 (161) | 478.56 (167) | 0.45 | 493.83 (167) | 956.79 (159) |
| Maravilha | IVB | GG | 0.64 | 709.30 (162) | 645.25 (159) | 0.62 | 686.49 (159) | 948.43 (160) |
| Honduras | LRC | GG | 0.63 | 695.03 (163) | 618.78 (162) | 0.56 | 616.60 (164) | 929.12 (164) |
| N.7384 | IVC | GG | 0.62 | 687.50 (164) | 630.57 (161) | 0.59 | 652.54 (161) | 933.17 (163) |
| Guapinha | LRC | GG | 0.57 | 630.42 (165) | 611.69 (164) | 0.57 | 627.22 (163) | 908.21 (165) |
| Vermelhão | LRC | GG | 0.55 | 612.78 (166) | 579.48 (165) | 0.52 | 579.50 (165) | 895.79 (166) |
| Yn1906-Uul 65 | IVC | GG | 0.52 | 576.95 (167) | 541.75 (166) | 0.49 | 537.05 (166) | 879.76 (167) |
| Tb47h-Mr-11-51-3 | IVC | GG | 0.49 | 543.21 (168) | 328.68 (168) | 0.29 | 323.39 (168) | 858.58 (168) |
Table 5: Estimates of adaptability (RPGV), stability (HMGV) and harmonic mean for adaptability and stability (HMRPGV) of the 20 most productive (20+) and 20 least productive accessions (20-), obtained from the joint analysis (BLUPj). The values between parenthesis are related to the ranking of the accessions according RPGV*GA. GA, yield general average.
Discussion
One of the great challenges of molecular genomics studies is to transfer the results to the routine practice of breeding programmes. The use of markers for assisted selection aims to accelerate the process of development of lines by prior identification of those lines possessing the favourable allele of genes of interest or markers linked to those genes of interest. This would increase the efficiency of breeding programmes because materials with a low potential for success in the programme would not be maintained in the experiments. Specifically, for the drought-tolerance trait, an initial screening that results in the reduction in the number of evaluated genotypes represents a significant saving of resources. This type of experiment is difficult to conduct due to the need for constant monitoring of the water-deficit conditions so that the deficit is high enough to kill the plants or low enough to prevent discrimination between the tolerant and susceptible plants. The translation of these markers to routine breeding based on marker assisted selection is difficult, because these traits are controlled for many loci that individually explain a small percentage of the phenotypic variation and the markers might be specific to a genetic background distinct from those evaluated or in the selection for environments different from those used in the experiments.
This work began with a gene selection, which were used to sequence 175 upland rice accessions. This set of accessions was evaluated at the Drought Phenotyping Experimental Station in Porangatu for three years (2010 to 2012). Considering the 276 SNPs obtained from the 32 sequenced genes, three were consistently associated with the 20 most productive accessions (SNP pattern AA) and the 20 least productive accessions (SNP patterns GG for S2, TT for S8 and CC for S9) in the 2012 experiment. The fact that the three genes located on chromosomes 2, 8, and 9 have the AA pattern related to higher productivity in the drought could have occurred due to chance, or somehow, being in linkage disequilibrium. This should be investigated in the future. According to prioritization (RiceNet V2; Lee et al., 2015) and co-expression network analyses (RiceFrend; Sato et al., 2013), these three genes are not connected. The gene LOC_Os02g09650 has an AP2 protein domain, and the ontology gene analysis shows that it has transcription factor activity (GO: 0003700), signal transduction (GO: 0007165), response to stress (GO: 0006950) and response to abiotic stimulus (GO: 0009628). In its gene model, it has only one exon and one transcript. This gene has been described as having a direct role in the response to drought (Dietz et al., 2010; Kazan, 2015). The gene LOC_Os08g13840 is the transcription factor WRKY25 (GO: 0003700 and GO: 0006355), and its gene model has two transcripts and two exons. This transcription factor has previously been associated with the response to drought by Tripathi et al. (2014). The action of transcription factors in response to drought has been well documented (Singh and Laxmi, 2015; Samad et al., 2017). The gene LOC_Os09g21120 (OsPUB6) is associated with catalysis of the transfer of ubiquitin from one protein to another (GO: 0004842), interacting selectively and non-covalently with any protein or protein complex (GO: 0005515) and the process in which one or more ubiquitin groups are added to a protein (GO: 0016567); furthermore, its gene model has only one exon and one transcript. Ubiquitination is a major type of post- translational modification of proteins that occurs in eukaryotic cells, and a cell death assay, in which one of the U-box genes was silenced, caused cell death in rice protoplasts, suggesting that ubiquitination might be involved in rice cell defence responses (Zeng et al., 2008).
Of the three genes associated with drought tolerance or susceptibility, two of these were identified by GWAS (LOC_Os02g09650, SNP S2; and LOC_08g13840, SNP S8), whereas one was identified by RNA-Seq (LOC_Os09g21120, SNP S9). The advantage of the GWAS analysis was that it was performed with phenotypic data obtained in field experiments, which indicates that such genes would be expressed, at least theoretically, in conditions similar to those found in crop cultivation. On the other hand, the operational cost to obtain the phenotypic data in the field of a given study population is much higher than that required to establish an RNA-Seq experiment in a greenhouse because field trials must be conducted at a phenotyping site where rainfall is prevented. In addition, the genotyping of tens to hundreds of accessions is costly. The analysis by RNA-Seq allows few accessions (even a single accession) to be evaluated in a greenhouse experiment, which allows more precise control of the water deficit. However, to identify the SNPs in these genes, a second round of analysis is necessary, such as by Capture-Seq approach used in this work. The SNPs related to drought tolerance obtained by RNA-Seq are identified in the transcribed genes, whereas the SNPs derived from a structural genome sequencing approach (such as genotyping by sequencing, or GBS) can be located both in genes and in intergenic sequences. The SNP 7, for example, selected to compose a marker panel for assisted selection for drought in rice (Pantalião et al., 2016), lies in an intergenic region, allowing a more comprehensive exploration of the genetic variability of structural DNA than the functional genome of rice.
As the SNPs derived from the three genes showed the same genotyping pattern (AA for the most productive genotypes and GG, TT and CC for the least productive genotypes under drought conditions), any of these three SNPs could be used in marker-assisted selection, with S2 being the one selected for this study.
Phenotypic analysis × genotypic analysis for drought tolerance
The genotype interaction per year was quite significant for grain yield. Of the 168 accessions analysed in all three experiments, 20 had above-average yield, while 35 were below average; that is, the remaining 113 accessions, or 67%, presented different productive performance in the three years of experiments. The comparative analysis of the AA pattern with the GG pattern of S2 marker showed that the most efficient way to perform the assisted selection was to select for the GG pattern of genotypes because it corresponded to 19 of the 20 least adapted genotypes in the joint analysis.
No distinction in adaptability was observed between the LRCs and improved varieties. That is, the breeder may prioritize the drought-tolerant materials derived from the breeding programme, as those materials already have other desirable phenotypes, such as plant architecture, cycle and grain quality. In contrast, when choosing landraces, additional work would be required to ensure that the derived inbred lines would have the favourable phenotype, due to the linkage between drought tolerance and undesirable traits (Vikram et al., 2015).
As drought tolerance is a complex trait, the identification of markers robust enough for use in assisted selection that can be used in multiple environments over several years and several populations is a difficult task (Kamoshita et al., 2008). The development of a technological product with these characteristics is difficult because of the different responses of the genotypes to the different environmental conditions that occur each year and at each site. The proposal to use the SNP S2 for the breeding programme is different, as this marker is based on the identification of the accessions that have the SNP GG pattern, which was mostly associated with accessions with grain yield below the experiments average. This would allow the selection of accessions with genotype AA, which, in the present study, represented 33.3% of the accessions evaluated. Fewer accessions (but with potential higher yield average under drought) would allow experiments with a greater number of replications to be performed, in at least three years, for a more consistent evaluation of the genotypes. This is a key point in regard to studying drought tolerance: the experiment must be repeated as often as possible to adequately estimate the productive potential of each genotype when grown under water deficit conditions. When selecting for a complex trait, such as drought tolerance, the following is important to consider: achieving balance between the size of the trial, the number of repetitions within the experiment, the number of years and the number of experimental sites. This decision should also consider the cost of implementing these trials.
Based on the results obtained in this work, SNP 2 assisted selection should be performed in a wide set of accessions and the selected ones would be evaluated in experiments with more repetitions for each location and for more than one experimental season. The removal of the accessions with the GG SNP 2 pattern could eliminate some of the most productive genotypes under drought conditions. However, considering the availability of drought tolerant materials in rice gene banks, the impact of the elimination of some of these accessions is reduced due to the discard of the clear majority of consistently drought susceptible accessions.
Conclusion
1) Both the RNA-Seq and GWAS methodologies were efficient in identifying SNP markers associated with drought tolerance or susceptibility. RNA-Seq is faster in identifying genes associated with the trait but requires a subsequent step to identify SNPs in these genes. GWAS requires the genotyping of many accessions and field experiments, but directly identifies SNP markers of both genes and intergenic regions.
2) The SNP markers derived from candidate genes obtained in the literature evaluated in this study were not useful for marker-assisted selection because they did not discriminate between the drought-tolerant and drought-susceptible individuals.
3) Of the 32 genes used in the identification of drought-tolerant genotypes, three could identify the susceptible genotypes consistently.
4) This work showed that at least three experiments per target site are required for genetic improvement for drought tolerance, regardless of the experimental design used.
5) The use of only one SNP marker means a significant reduction in the cost of genotyping, allowing many genotypes to undergo this screening. The selected accessions can then be used to conduct smaller, and consequently cheaper and more precise, experiments.
Acknowledgments
The Coordination for the Improvement of Higher Education Personnel/Ministry of Education (CAPES/MEC) for the grants to AFV; National Council for Scientific and Technological Development (CNPq) for the grants to CB and RPV and the Brazilian Agricultural Research Corporation Rice and Beans (EMBRAPA Rice and Beans) for support for this research.
About the Authors
Corresponding Author
Claudio Brondani
Embrapa Arroz e Feijão, CP 179, CEP 74001-970, Goiânia – GO, Brasil
- Email:
- claudio.brondani@embrapa.br
References
- Abadie T, Cordeiro CMT, Fonseca JR, Alves RBN, et al. (2005). Construção de uma coleção nuclear de arroz para o Brasil. Pesqui. Agropecu. Bras. 40: 129-136.
- Abreu FRM, De Deus KE, Pereira WJ, Silveira RDD, et al. (2016). Expression of rice genes homologous of Arabidopsis genes previously related to drought tolerance. A.J.C.S. 10: 1266-1272.
- Babu RC (2010). Breeding for drought resistance in rice: an integrated view from physiology to genomics. Electron. J. Plant Breed. 1: 1133-1141.
- Barbosa Filho MP, Yamada T (2002). Upland rice production in Brazil. Better Crops International 16: 43-46.
- Bentley DR, Balasubramanian S, Swerdlow HP (2008). Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53-59. 10.1038/nature07517.
- Bernier J. Kumar A, Venuprasad R, Spaner D, et al. (2009). Characterization of the effect of a QTL for drought resistance in rice, qtl12.1, over a range of environments in the Philippines and eastern India. Euphytica 166: 207-217. 10.1007/s10681-008-9826-y.
- Borah P, Sharma E, Kaur A, Chandel G, et al. (2017). Analysis of drought-responsive signalling network in two contrasting rice cultivars using transcriptome-based approach. Sci. Rep. 7: 42131. 10.1038/srep42131.
- Counce PA, Keislingb TC, Mitchellb AJ (2000). A uniform, objective, and adaptive system for expressing rice development. Crop Science 40: 436-443.
- Degenkolbe T, Do PT, Kopka J, Zuther E, et al. (2013). Identification of drought tolerance markers in a diverse population of rice cultivars by expression and metabolite profiling. PLoS ONE 8: e63637. 10.1371/journal.pone.0063637.
- Dietz KJ, Vogel MO, Viehhauser A (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signaling. Protoplasma 245: 3-14. 10.1007/s00709-010-0142-8.
- Dixit S, Huang BE, Sta Cruz MT, Maturan PT, Ontoy JCE et al. (2014). QTLs for tolerance of drought and breeding for tolerance of abiotic and biotic stress: an integrated approach. PLoS ONE 9: e109574. 10.1371/journal.pone.0109574.
- Fageria NK, Wander AE, Silva SC (2014). Rice (Oryza sativa) cultivation in Brazil. Indian J. Agron. 59: 350-358.
- Fedoroff NV, Battisti DS, Beachy RN (2010). Radically rethinking agriculture for the 21st century. Science 327: 833-834. 10.1126/science.1186834.
- Heinemann AB, Barrios-Perez C, Ramirez-Villegas J, Arango-Londoño D, et al. (2015). Variation and impact of drought-stress patterns across upland rice target population of environments in Brazil. J. Exp. Bot. 66: 3625-3638. 10.1093/jxb/erv126.
- Huang L, Zhang F, Zhang F, Wang W, et al. (2014). Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC Genomics 15: 1026. 0.1186/1471-2164-15-1026.
- Jain M, Moharana KC, Shankar R, Kumari R, et al. (2014). Genomewide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance. Plant Biotechnol. J 12: 253-264. 10.1111/pbi.12133
- Kamoshita A, Babu RC, Boopathi NM, Fukai S (2008). Phenotypic and genotypic analysis of drought-resistance traits for development of rice cultivars adapted to rainfed environments. Field Crops Res. 109: 1-23. 10.1016/j.fcr.2008.06.010.
- Kazan, K (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20: 219-229. 10.1016/j.tplants.2015.02.001.
- Kumar A, Dixit S, Ram T, Yadaw RB, et al. (2014). Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J. Exp. Bot. 65: 6265-6278. 10.1093/jxb/eru363.
- Lee T, Oh T, Yang S, Shin J, et al. (2015). RiceNet v2: an improved network prioritization server for rice genes. Nucleic Acids Res. 43: 122-127. 10.1093/nar/gkv253.
- Neves LG, Davis JM, Barbazuk WB, Kirst M (2013). Whole-exome targeted sequencing of the uncharacterized pine genome. Plant J 75: 146-156. 10.1111/tpj.12193.
- Neves LG, Davis JM, Barbazuk WB, Kirst M (2014). A high-density gene map of loblolly pine (Pinus taeda L.) based on exome sequence. G3 4: 29-37. 10.1534/g3.113.008714.
- Pantalião GF, Narciso M, Guimarães C, Castro A, et al. (2016). Genome wide association study (GWAS) for grain yield in rice cultivated under water deficit. Genetica 144: 651-664. 10.1007/s10709-016-9932-z.
- Parida SK, Mukerji M, Singh AK, Singh NK, et al. (2012). SNPs in stress-responsive rice genes: validation, genotyping, functional relevance and population structure. BMC Genomics 13: 426. 10.1186/1471-2164-13-426.
- Patterson HD, Thompson R (1971). Recovery of inter-block information when block sizes are unequal. Biometrika 58: 545-554.
- Resende MDV (2004). Métodos estatísticos ótimos na análise de experimentos de campo. Embrapa Florestas, Colombo.
- Resende MDV, Duarte JB (2007). Precisão e controle de qualidade em experimentos de avaliação de cultivares. Pesq. Agropec. Trop. 37: 182-194.
- Samad AFA, Sajad M, Nazaruddin N, Fauzi IA, et al. (2017). MicroRNA and transcription factor: key players in plant regulatory network. Front. Plant. Sci. 8: a565. 10.3389/fpls.2017.00565.
- Sato Y, Namiki N, Takehisa H (2013). RiceFREND: A platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res 41: 1214-1221. 10.1093/nar/gks1122.
- Serraj R, Kumar A, McNally KL, Slamet-Loedin I, et al. (2009). Improvement of drought resistance in rice. Adv. Agron. 103: 41-99. 10.1016/S0065-2113(09)03002-8.
- Serraj R, McNally KL, Slamet-Loedin I, Kohli A, et al. (2011). Drought resistance improvement in rice: an integrated genetic and resource management strategy. Plant Prod. Sci. 14: 1-14. 10.1626/pps.14.1.
- Shaar-Moshe L, Hübner S, Peleg Z (2015). Identification of conserved drought-adaptive genes using a cross-species meta-analysis approach. BMC Plant Biology 15: 111. 10.1186/s12870-015-0493-6.
- Silveira RDD, Abreu FRM, Mamidi S, McClean PE, et al. (2015). Expression of drought tolerance genes in tropical upland rice cultivars (Oryza sativa). Genet. Mol. Res. 14: 8181-8200.
- Singh D, Laxmi A (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6: a895. 10.3389/fpls.2015.00895.
- Swamy BPM, Kumar A (2013). Genomics-based precision breeding approaches to improve drought tolerance in rice. Biotechnol Adv 31: 1308-1318. 10.1016/j.biotechadv.2013.05.004.
- Swamy M, Ahmed HU, Henry A (2013). Genetic, physiological, and gene expression analyses reveal that multiple qtl enhance yield of rice mega-variety IR64 under drought. PLoS ONE 8: e62795. 10.1371/journal.pone.0062795.
- Tripathi P, Rabara RC, Rushton PJ (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239: 255-266. 10.1007/s00425-013-1985-y.
- Vikram P, Swamy BPM, Dixit S, Singh R, et al. (2015). Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci. Rep. 5: 14799. 10.1038/srep14799.
- Zeng LR, Park CH, Venu RC, Gough J, et al. (2008). Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 1: 800-815. 10.1093/mp/ssn044.
Keywords:
Download:
Full PDF- Share This