Identification of Klf6-Related Super Enhancer in Human Hepatoma (HepG2) Cells by CRISPR Technique
Received: September 11, 2017
Accepted: November 08, 2017
Published: November 29, 2017
Genet.Mol.Res. 16(4): gmr16039841
DOI: 10.4238/gmr16039841
Abstract
The Klf6 gene is a tumor suppressor gene belonging to the family of the Klf gene and closely associated with tumor formation. Recently, super enhancers(SEs) have been shown to play a particularly important role in regulating cell identity and are attempting to evaluate the in vivo function of SEs. But direct functional evidence of SE associated with Klf6 is lacking. Using genomic editing technology, we have attempted to identify super enhancer associated with the expression of Klf6 and clarify its function in human hepatoma (HepG2) cells. As a result, it identified the Klf6 related SE and demonstrated that the Klf6-related SE is responsible for more than 80% of Klf6 gene expression. It also revealed the hierarchical structure of the Klf-6 related SE and the function of individual enhancers. Our results provide the functional significance of the super enhancer in understanding the transcriptional regulation mechanism of Klf6
Introduction
The Klf6 gene is a tumor suppressor gene belonging to the family of the Klf. It is involved in the physiological processes such as cell differentiation and development, cell growth signal transduction, cell proliferation, apoptosis, and angiogenesis [Michaela et al,2006; A.C. Racca et al,2016; Magali et al,2011]. Inhibition of Klf6 gene expression in tumor cells is closely related to tumor formation and development. The Klf6 gene is expressed in several tumours, but its degree of expression is mutually independent [Ricado et al, 2011].
Recently, mutation of the Klf6 gene has been observed in various tumours including liver cancer, stomach cancer, and colon cancer [Ceshi et al, 2003; Magali et al,2011; Sigal et al, 2007]. As a result of mutation, it has been shown to induce the tumor. This suggests that identifying the transcriptional regulatory mechanism of the klf6 gene is crucial in understanding tumorigenesis [ Jaya et al, 2009; Jaya et al, 2009; Analisa et al, 2009].
Enhancer plays a very important role in regulating gene transcription in multicellular animals. Recent genome-wide analyses have revealed that enhancer is very abundant in the genome, and these represent the cell identity in a way that regulates their distinctive transcriptional programs in individual cells [Jonathan et al, 2014; Andrienne et al, 2015; Creyghton et al, 2010]. Recently it has been mentioned as a direct functional evidence that super enhancers play a particularly important role in regulating cell identity and disease [Anna et al, 2014; Marc et al, 2014; Denes et al, 2015; Sophia et al, 2016; Denes et al, 2013].
It has been confirmed that the recently developed crispr/cas9 system can be used as a tool for evaluating enhancer functions in vivo with highly effective genome editing technology [Jialiang et al, 2016; Mali et al, 2013; Cong et al, 2013]. Recent studies have used the CRISPR technique to assess the importance of a SE near GATA2 gene in chronic myeloid leukemia, suggesting that SE is responsible for 80% of GATA2 gene expression [Stefan et al, 2014]. It is also shown that a SE of Sox2 in the mouse gemone is ~100 kb away from the Sox2 gene and is responsible for 90% of the Sox2 gene expression [Yan et al, 2014]. Experimental studies of the super enhancer are now being extensively undertaken, while direct functional evidence of SE associated with Klf6 is lacking.
Based on these developments, we attempted to find an SE associated with the expression of Klf6. It identified a super enhancer that are marked by activated histone markers in regions near Klf6 gene of human hepatoma (HepG2) cells. It demonstrated that the Klf6-related SE is responsible for more than 80% of Klf6 gene expression in hepatoma (HepG2) cells using a double- CRISPR excision method.
Materials and Methods
Cell culture and transfection experiments
Human HepG2 cells were purchased from the typical training content preservation committee cell bank, Chinese Academy of Sciences (China). The Cells were cultured as described previously [Michaela et al, 2006]. Importantly, the cells were passaged twice. The cells were transfected for 24 h before transfection. The cells were cultured with CRISPR plasmid and Lipofectamine 2000 (Addgene) for 6 h. After transfection, the cells were cultured for 48 h in fresh medium, 0.5 μg/mL of puromycin (Sigma) was supplemented to the cell culture medium for 3 days to select for the transfected cells. Cells were cultured for 24 h in a fresh medium, and clones were obtained by limiting dilution method, from which target clones were selected.
Design of CRISPR constructs
The target-specific CRISPR guide RNAs (sgRNAs) were designed using an online tool (http://crispr.mit.edu/). sgRNAs were designed to optimize uniqueness and to have limited off-target. The designed oligonucleotides were inserted into pSpCas9 (BB)-2A-GFP (PX458) plasmid (Addgene) and used for cell transfection experiments. The characteristics of the designed sgRNA-specifying oligo sequences are shown in Table 1.
| SgRNA-A | SgRNA-B | ||||||
|---|---|---|---|---|---|---|---|
| No | Deletion(kb) | Chr | Genomic Coordinates | Sequence | Genomic Coordinates | Sequence | |
| 1 | E | 29 | 10 | 3,873,065-3,873,082 | CATATGCTACATACCAGC | 3,844,061-3,844,078 | CATTTGACCAGCCGCCCG |
| 2 | E1 | 6.5 | 10 | 3,850,540-3,850,557 | AAGGGCAGGCTCCACGCC | 3,844,061-3,844,078 | CATTTGACCAGCCGCCCG |
| 3 | E2 | 6.7 | 10 | 3,850,540-3,850,557 | AAGGGCAGGCTCCACGCC | 3,857,219-3,857,236 | CGTGCAACGAGTGAATGG |
| 4 | E3 | 6.6 | 10 | 3,873,065-3,873,082 | CATATGCTACATACCAGC | 3,866,418-3,866,435 | GGATGCGACAGGGTCCTT |
| 5 | E1-2 | 13.2 | 10 | 3,857,219-3,857,236 | CGTGCAACGAGTGAATGG | 3,844,061-3,844,078 | CATTTGACCAGCCGCCCG |
Table 1: sgRNA localization, deletion size, chromosome, genomic coordinates (GRCh37/hg19), and sequence for each sgRNA pairs
PCR and Sanger sequencing for Super enhancer deletion clones
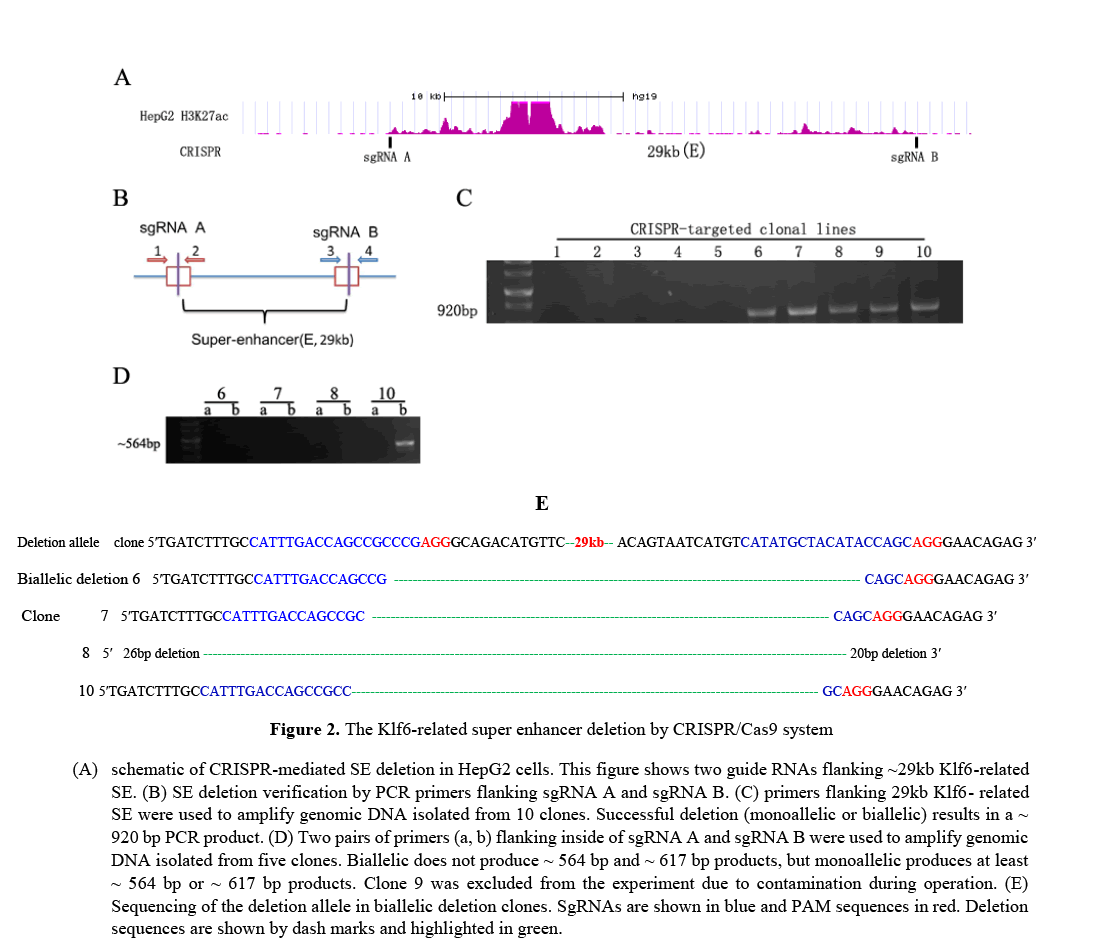
Genomic DNA was isolated from the super enhancer deletion (SE-del) clones using DNeasy Blood & Tissue Kit (Qiagen). To select the deleted clones, the primer flanking the outside of CRISPR sgRNAs were designed and amplified a ~ 29kb region. The size of the amplified product is ~ 920 bp and the primer is 5'- GCCTGAGATGAGAGTTACAG-3'(forward) and 5'-CTGTACGTTTCGGAAAGT TG-3' (reverse). The PCR condition: after initial incubation at 95 ° C for 5 minutes, a total of 35 cycles were performed, each including 94 ° C for 30 seconds, 55 ° C for 40 seconds, and 72 ° C for 1 min.
In order to evaluate the biallelic or monoallelic deletion characteristics of the target clones, two pairs of primers were designed to flank the inside of the CRISPR sgRNAs and amplified ~ 564 bp and ~ 617 bp products. It was verified by sanger sequencing.
Real-time quantitative PCR
Total RNA was isolated from the target clones using TRIzol (Life Tech) extraction method. A total of 1 � RNA was used as a template for cDNA synthesis. cDNA (100ng) was used as a template for PCR amplification and SYBR green-based QRT-PCR was performed on an ABI PRISM 7900HT Sequence Detection System.
Primer sequences for the Klf6 genes and beta-actin are as follows: 5′-CGGACGCA CACAGGAGAAAA-3′/5′-CGGTGTGCTTTCGGAAGTG-3′ (KLF6) and 5′-GAAATCGTGCGTG ACATTAAG-3′/5′-CTAGAAGCATTTGCGGTGGACGATGAGGGGCC-3′ (beta -actin). A total of 40 cycles were performed, each including 95°C for 30 seconds, 54°C for 30 seconds, and 72°C for 30 seconds. Assays were done in triplicate and Relative levels of gene expression were normalized to beta-actin.
Western blot assay
Western blot assays were performed as previously described [Michaela et al, 2006]. The following antibodies were used: rabbit polyclonal antibody to Klf6 (R-173): goat polyclonal to beta-actin (I-19). Visualisation was performed by the enhanced chemoluminescent method (Amersham International, Freiburg, Germany) and quantified with the BIOQUANT NOVA imaging system (BIOQUANT NOVA PRIME Measurement Software).
Chromosome Conformation Capture
Chromosome conformation capture were performed as previously described [N. naumova et al, 2012]. The cells were cross-linked with 1% formaldehyde in the presence of 10% FBS for 10 min at room temperature and lysed on ice. Collected nuclei were digested with 400 U of HindIII restriction enzyme over night at 37 °C with shaking. Ligation was performed for 4 h at 16 °C and 30min at room temp with 100 U of T4 DNA ligase. DNA was ethanol precipitated and analysed by PCR using primers designed to amplify across junctions of Klf6 promoter and each enhancer (E1, E2, E3). As a control (normal hepatocyte), genomic DNA regions covering restriction sites of interest was purified and analysed by PCR using the same primers.
Results
Identification of Klf6-related super enhancers in various human cancer cell lines
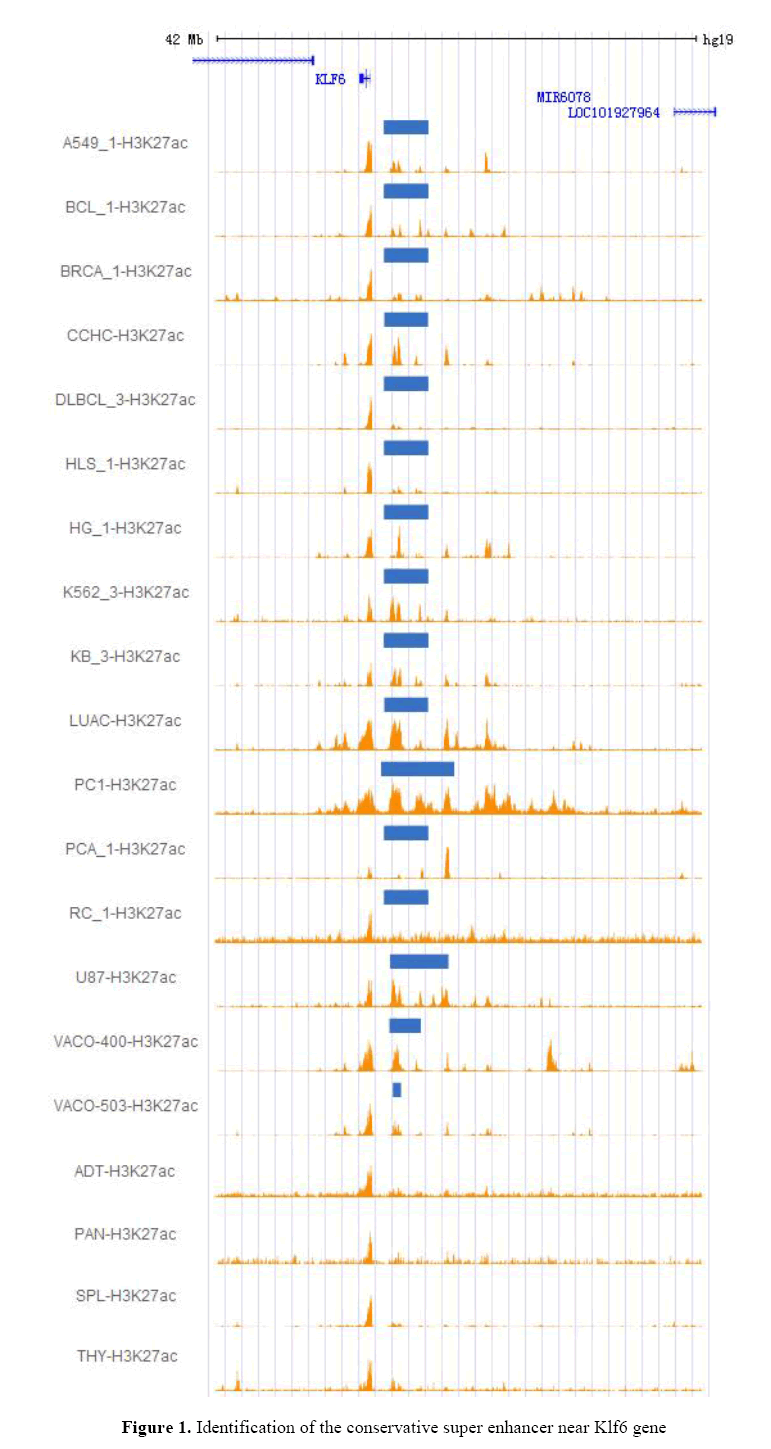
Using CHIP-seq data, it confirmed that a super-enhancer is commonly present in upstream of the Klf6 gene of 16 cancer cell lines [Yanjun et al, 2016]. As a result, it observed a super enhancer conservatively present in 16 cancer cell lines, approximately ~26 kb away from the Klf6 gene (Figure 1). As shown in Figure 1, the super enhancer was not observed in adipose, pancreas, spleen, and thymus tissues, unlike in selected cancer cell lines. For example, panc1 has SE but does not exist in normal pancreas. These results show that the super enhancer can be formed by the changes in upstream regions of Klf6 before and after tumor formation. This hypothesis is supported by the literature [Jonathan et al,2014]. This contains a relatively large ~29kb sequence.
The figure shows 20 tracks of H3k27ac. The upper 16 tracks show an conservative super enhancer on various cancer cell lines, and the lower 4 tracks have not super enhancer as contrast. The blue bands represent the super enhancer. These data were obtained from Super Enhancer Archive (SEA, http://sea.edbc.org). Based on this, it selected human HepG2 cells among 16 human cancer cell lines and verified their functional characteristics, assuming that this super enhancer is associated with the expression of Klf6 gene in human HepG2 cells.
CRISPR/Cas9 mediated deletion analysis of the Klf6-related super enhancer
To elucidate the functional properties of the Klf6-related super enhancer, it selected a human HepG2 cells and used CRISPR/Cas9 technology to eliminate the enhancer sequence predicted as a super enhancer. Recently, the CRISPR/Cas9 genome editing technology has been regarded as one of the simplest and most effective methods to delete intron as well as exon. We used this editing technique to obtain 10 HepG2 cell clones. One of 10 clones were a monoallelic clone and 3clones were biallelic clones, as confirmed by PCR analysis and sanger sequencing (Figure 2). As shown in Figure 2.C and Figure 2.D, our experimental results show that double CRISPR/Cas9 genome editing technology can eliminate large genome sequences with very high efficiency.
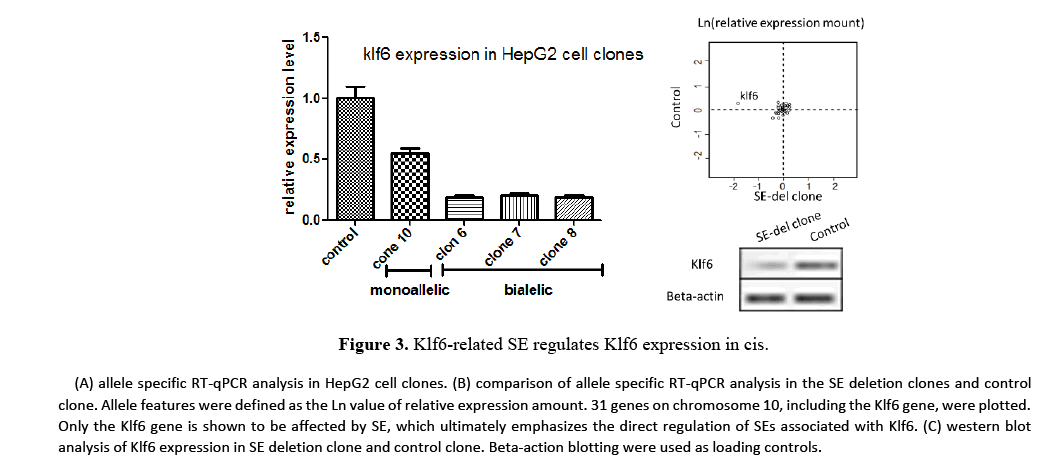
RT-qPCR analysis of the Klf6-related SE
It performed allele-specific RT-qPCR analysis on HepG2 cell clones to quantify the effect of the Klf-related SE on Klf6 gene transcriptions (Figure 3). In monoallelic SE deletion clone, Klk6 expression decreased to approximately 50% of total Klf6 expression, while in biallelic SE deletion clones, Klf6 expression level decreased by more than 80%. These results show that a single SE regulates transcription of the Klf6 gene in cis. It performed RT-qPCR analysis on a total of 30 upstream and downstream genes, each 3 Mb apart from the Klf6 gene, to further assess the cis effect of SE deletion on transcription. These results also emphasized that the Klf6-related SE is required for the transcription of the Klf6 gene. Importantly, there is no effect on the expression of genes present on the same chromosomes (~6 Mb region) after deletion of the Klf6 related SE. This suggests that the Klf6 gene may be a target gene for the Klf6 related SE.
It next performed western blot analysis to assess the expression level of Klf6 on SE deletion clone (Figure 3.C). Western blot analysis was also significantly lower than that of control in the SE-del clone as in RT-qPCR analysis. These results show that the expression of Klf6 is regulated by the Klf6-related super enhancer.
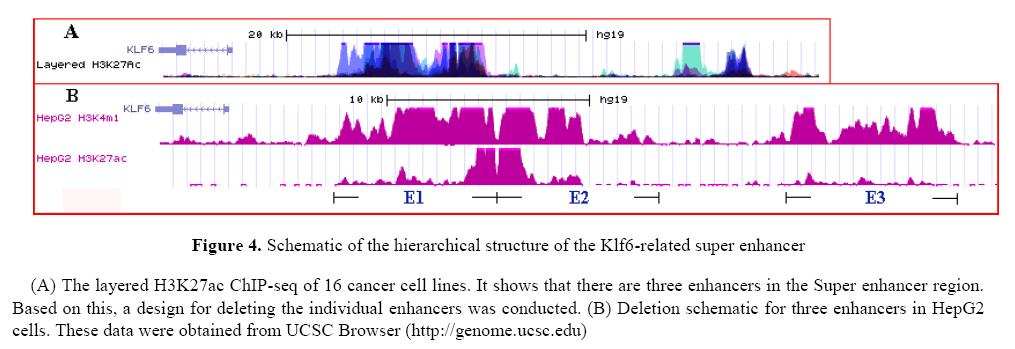
The hierarchical structure of the Klf6-related super enhancer and RT-qPCR analysis
Based on the H3K4me1 and H3K27ac ChIP-seq it divided the super enhancer sequence into three enhancers and designed sgRNAs that flank the individual enhancers (see method). The Figure 4 show the hierarchical structure of the Klf6-related super enhancer and deletion schematic for three enhancers in HepG2 cells. Based on this results, it obtained clones containing biallelic deletion of each enhancer (E1, E2, E3, E1-2).
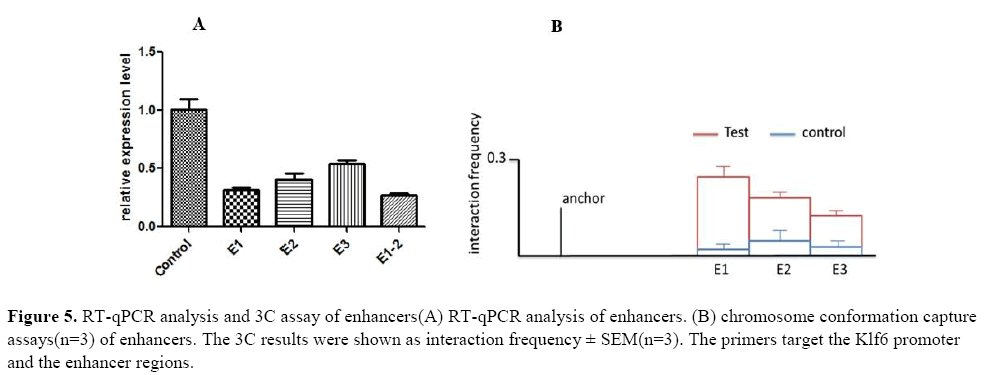
Next, we isolated RNA from clones and used RT-qPCR analysis to evaluate the effect of individual enhancers on the expression of Klf6 (Figure 5.A). As shown in the figure, the deletion of the individual enhancers surprisingly had a large effect on the expression of Klf6. Among them, the expression of Klf6 was greatly reduced in the E1-del clone and the expression of the E3-del clone was the smallest.
These results suggest that the enhancers far from the Klf6 gene are less likely to regulate the expression of the Klf6 gene. Eventually, the E1 enhancer shows that it is functionally stronger than its neighboring enhancers in regulating transcriptional activation.
Thus, our enhancer deletion assays demonstrate that the Klf6-related SE is composed of a functional hierarchy containing components that modulate the transcriptional activation of Klf6.
Next, it performed chromosome conformation capture assay to elucidate the physical interaction between the enhancers and the Klf6 promoter (Figure 5.B). This result highlight that the Klf6-related SE regulates Klf6 gene through long-range DNA looping.
Discussion
We identified and the Klf6-related super enhancer that regulates Klf6 expression in human HepG2 cells. The Klf6-related super enhancer we identified is a H3K27ac-rich intronic region (~ 29kb) that fits the recently defined super enhancer criteria. Recently, several studies have suggested that super enhancers play an important role in maintaining and regulating cell identity in various tumor cells and stem cells, and have also attempted to establish methods of drug development and treatment targeting these SEs [Jakob et al, 2013]. Our experimental results are merely functional study data of SE supporting the experimental direction of super enhancers.
It observed that an Klf6-related super enhancer present in 16 cancer cells was not present in normal cells (Figure 1). This suggests that the Klf6-related super enhancer can be formed in the posterior and posterior processes of tumor formation in normal cells. Some researchers [Emily et al, 2015] have found evidence of an epigenetic basis for the transition between WDLPS (well differentiated LPS) and DDLPS (dedifferentiated LPS) in Liposarcoma (LPS). They observed elevated H3K9me3 levels in DDLPS tumours and found that increased H3K9me3 may mediate through Klf6 and these increased H3K9me3 sites are associated with super enhancers as the upstream region of Klf6. These experimental data support our hypothesis on the formation of the Klf6-related super enhancer [Jonathan et al, 2014]. Based on these studies, it would be very interesting to further study the process of tumor formation with the super enhancer.
Next, it successfully deleted ~29 kb non-coding region using the CRISPR/Cas9-mediated genomic deletion method. It found that the deletion of the Klf6-related super enhancer reduced by more than 80% of the Klf6 expression and did not affect the expression of other neighboring genes (Figure 3). This suggests that the Klf6-related super enhancer modulates the expression of Klf6 in cis.
In addition, with the remarkable effectiveness of the CRISPR/Cas9 system, it could further elaborate the hierarchy of the Klf6-related super enhancer. Based on H3K4me1 and H3K27ac ChIP-seq, it identified 3 enhancers in the Klf6-related super enhancer and found that these genes also functioned to regulate the expression of Klf6. As a result, it was confirmed that the Klf6-related super enhancer was composed of 3 enhancers and regulated the expression of Klf6 (Figure 5).
In addition, chromosome conformation capture assay (3C) analysis revealed that Klf6-related SE regulates the Klf6 gene through long-range DNA looping. finally, it believes that the CRISPR / Cas9 system can be a powerful tool in studying in vivo super enhancer function. There are several problems to be solved the function of the Klf6-related super enhancer in the future. What are the transcription factors involved in the expression of Klf6, and what are its functional interactions? Resolving these problems will be very robust in understanding the function of super enhancer and applying it to future applications.
Acknowledgments
We thank Ph.D. wuqiong for guidance in this search. This work was supported by the Program for Chang jiang Scholars and Innovative Research Team in University (IRT1220) and the National Natural Science Foundation of China (Grant No. 54289562).
Conflicts of interest
The authors declare that they have no conflict of interest.
About the Authors
Corresponding Author
Kum Chol Ri
School of Management, Harbin Institute of Technology, China
- Email:
- 15124579835@163.com
References
- AC Racca, ME Ridano, CL Bandeira, E Bevilacqua (2016). Low oxygen tension induces Krüppel-Like Factor 6 expression in trophoblast cells. Placenta. 45: 50-57. https://doi.org/10.1016/j.placenta.2016.07.006
- Adrienne R, Niederriter, Arushi Varshney, Stephen C. J. Parker (2015). Super Enhancers in Cancers, Complex Disease, and Developmental Disorders. Genes. 6:1183-1200. https://doi.org/10.3390/genes6041183
- Analisa DiFeo, John A Martignetti, Goutham Narla (2009). The role of KLF6 and its splice variants in cancer therapy. Drug Resistance Updates 12: 1-7. https://doi.org/10.1016/j.drup.2008.11.001
- Anna Vaharautio, Jussi Taipale (2014). Cancer by super-enhancer. Science. 346(6215)1291-1292. https://doi.org/10.1126/science.aaa3247
- Ceshi Chen, Eija-Riitta Hyytinen, Xiaodong Sun, Heikki J (2003). Mutation, and Loss of Expression of KLF6 in Human Prostate Cancer. American Journal of Pathology. 162:1349-1354.
- Cong L, Ran FA, Cox D, Lin S (2013) et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 339: 819-823.
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 107: 21931-21936. https://doi.org/10.1073/pnas.1016071107
- Denes Hnisz, Brian J. Abraham, Tong Ihn Lee, Ashley Lau, et al. (2013). Super-Enhancers in the Control of Cell Identity and Disease. Cell, 155, 934–947. https://doi.org/10.1016/j.cell.2013.09.053
- Denes Hnisz, Jurian Schuijers, Charles Y. Lin, Abraham S. Weintraub (2015). Young, Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super- Enhancers. Molecular Cell, 58: 1-9.
- Emily Z Keung, Kadir C, Akdemir, Ghadah AA, et al. (2015). Increased H3K9me3 drives dedifferentiated phenotype via KLF6 repression in liposarcoma. J Clin Invest. 125(8):2965-2978. https://doi.org/10.1172/jci77976
- Jakob Love´n, Heather A Hoke, Charles Y Lin, et al. (2013). Selective Inhibition of Tumor Oncogenes by Disruption of Super Enhancers. Cell, 153: 320-334. https://doi.org/10.1016/j.cell.2013.03.036
- Jaya Sangodkar, Analisa DiFeo, Lauren Feld, Romina Bromberg, et al. (2009). Targeted reduction of KLF6-SV1 restores chemotherapy sensitivity in resistant lung adenocarcinoma. Lung Cancer, 66:292-297. https://doi.org/10.1016/j.lungcan.2009.02.014
- Jaya Sangodkar, Jiayan Shi, Analisa DiFeo, Rachel Schwartz, et al. (2009). Functional role of the KLF6 tumour suppressor gene in gastric cancer. European Journal of Cancer. 45: 666-676.https://doi.org/10.1016/j.ejca.2008.11.009
- Jialiang Huang, Xin Liu, Dan Li (2016). Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Developmental Cell. 36: 9-23.https://doi.org/10.1016/j.devcel.2015.12.014
- Jonathan D Brown, Charles Y Lin, Qiong Duan, Gabriel Griffin, et al. (2014). NF-kB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Molecular Cell 56:1-13. https://doi.org/10.1016/j.molcel.2014.08.024
- Magali Humbert, Veronika Halter, Deborah Shan, Judith Laedrach, et al. (2011). Deregulated expression of Kruppel-like factors in acute myeloid leukemia. Leukemia Research. 35: 909-913. https://doi.org/10.1016/j.leukres.2011.03.010
- Mali P, Yang L, et al. (2013). RNA-guided human genome engineering via Cas9. Science. 339(6121): 823-826.https://doi.org/10.1126/science.1232033
- Marc R Mansour, Brian J Abraham, Lars Anders, Alla Berezovskaya, et al. (2014). An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science, 346: 6215.https://doi.org/10.1126/science.1259037
- Michaela S Banck, Simon W Beaven, Goutham Narla, Martin J Walsh (2016). KLF6 degradation after apoptotic DNA damage. FEBS Letters. 580: 6981-6986. https://doi.org/10.1016/j.febslet.2006.10.077
- N Naumova, EM Smith, Y Zhan, J Dekker (2012). Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 58(3): 1-27. https://doi.org/10.1016/j.ymeth.2012.07.022
- Ricardo C Gehrau, Diego S D’Astolfo, Verónica Andreoli, José L Bocco (2011). Differential expression of the klf6 tumor suppressor gene upon cell damaging treatments in cancer cells. Mutation Research, 707: 15-23. https://doi.org/10.1016/j.mrfmmm.2010.12.002
- Sigal Kremer-Tal, Goutham Narla, Yingbei Chen, Eldad Hod, et al. (2007). Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. Journal of Hepatology, 46: 645-654. https://doi.org/10.1016/j.jhep.2006.10.012
- Sophia Pinz, Samy Unser, Anne Rascle (2016). Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer. BMC Molecular Biol. 17:10. https://doi.org/10.1186/s12867-016-0063-y
- Stefan Groschel, Mathijs A Sanders, Remco Hoogenboezem, Elzo de Wit (2014). Single Oncogenic Enhancer Rearrangement Causes Concomitant EVI1 and GATA2 Deregulation in Leukemia. Cell. 157:369-381. https://doi.org/10.1016/j.cell.2014.02.019
- Yan Li, Choe M Rivera, Haruhiko Ishii, Fulai Jin (2015). CRISPR Reveals a Distal Super-Enhancer Required for Sox2 Expression in Mouse Embryonic Stem Cells. Plos one. 9(12), e114485. https://doi.org/10.1371/journal.pone.0114485
- Yanjun Wei, Shumei Zhang, Shipeng Shang, Bin Zhang, et al. (2016). SEA: a super-enhancer archive. Nucleic Acids Research. D172-D179. https://doi.org/10.1093/nar/gkv1243
Keywords:
Download:
Full PDF- Share This