Genetic diversity in natural populations of Stylosanthes scabra using ISSR markers
Received: November 14, 2017
Accepted: December 08, 2017
Published: January 11, 2018
Genet.Mol.Res. 17(1): gmr16039866
DOI: 10.4238/gmr16039866
Abstract
Genetic diversity is the basis for genetic improvement, providing genes for new cultivars. Stylosanthes scabra is a legume used in pastures, presenting economic importance in tropical and subtropical regions. Molecular markers are targeted in population studies, thus we evaluated the genetic diversity distribution of S. scabra in Brazilian semi-arid northeastern among and within populations, from molecular markers ISSR. Four natural populations of S. scabra located in the municipalities of Santa Cruz do Capibaribe, Floresta, Sertânia and Petrolina from Pernambuco state were used. We selected seven ISSR primers for amplification and analyzed 75 individuals. A total of 88 bands were amplified with a percentage of 95.27% polymorphism at the species level. AMOVA test revealed that 40.0% of the total genetic variation is within populations while 60.0% among populations. The population differentiation was 0.332 and the migrant’s number per generation was 0.5. Grouping analysis confirmed a high level of differentiation among populations. The populations with the greatest variability were Santa Cruz do Capibaribe and Petrolina, and the ISSR markers were efficient for the genetic diversity quantification in S. scabra, which presented greater variability among populations than within populations. In short, this variability will be very useful in future breeding programs for this species
Introduction
The scarcity of pastures in Brazilian semi-arid north-eastern in dry seasons has stimulated the advancement in studies of native forage plants (Santana Neto et al., 2015). Forage legumes can be used as a protein source to correct the deficiency of natural pastures (Tufarelli et al., 2010), moreover contain high levels of minerals and vitamins (Idowu et al., 2013).
The incorporation of forage legumes into the ruminant diet as a complementary food improves feed intake and feed converting (Pen et al., 2013). Thus, the inclusion of these legumes in animal feed also reduces the costs with the compound (Olafadehan et al., 2014). The genus Stylosanthes is a forage legume, which has extensive adaptation and resistance to biotic and abiotic factors (Pangga et al., 2004, Nagaich et al., 2013), with erect and prostrate species, predominantly perennial (Costa et al., 2008). This legume obtain nitrogen by association with diazotrophic bacteria (Mendonça et al., 2017), and can be indicated for the recovery of degraded pastures (Fabrice et al., 2015). However, Stylosanthes scabra can develop on soils with low levels of available nutrients, particularly phosphorus, conferring an advantage over other species (Gonzalez et al., 2000).
There are twenty-five species of Stylosanthes in Brazil and thirteen of them are found only in national territory (Santos-Garcia et al., 2011). S. scabra is a species with a large occurrence in South America (Calles and Schultze-Kraft, 2016), and this species is more common in the Brazilian Northeast, occurring in different environmental conditions (Oliveira et al., 2016). In other states from Brazil, this species occurs in Goiás, Minas Gerais, São Paulo (Brandão and Costa, 1979), Mato Grosso and Mato Grosso do Sul (Costa et al., 2008).
Stylosanthes scabra has some commercial cultivars, such as the cultivar Seca, launched in partnership between the International Center for Tropical Agriculture (CIAT) and the Commonwealth Scientific and Industrial Research Organization (CSIRO), which has stood out for its adaptability and agronomic performance under rainfed conditions in the subtropical climate. The cultivar Seca contains more than 17% of crude protein with low level of tannin (Mpanza et al., 2013). According to Akinlade et al. (2008), S. scabra can obtain a dry mass production up to 1.97 Ton.ha-1.
Exploring the full potential of S. scabra is essential to know the genetic variability about this species, supporting plant-breeding programs (Araújo et al., 2016). The genetic diversity evaluation of the species within and among populations can generate information for the in situ genetic conservation of the species natural populations (Gonçalves et al., 2010). In addition, the existence of variability in natural populations allows the evolution of new genetic combinations (Medrano et al., 2014), which may present greater capacity for evolution and adaptation to changes in environmental conditions (Srihari et al., 2013).
Four types of markers are used during the genetic diversity characterization: morphological, biochemical, molecular, and cytological. The molecular types present the particularity of use under analysis at any stage of plant development (Martuscello et al., 2015, Vieira et al., 2015). Among molecular markers, ISSRs (Inter Simple Sequence Repeat) stand out, because they do not require prior information of the DNA sequence and present laboratory procedures with good transferability (Dias et al., 2015). ISSR are semi-arbitrarily amplified by PCR (polymerase chain reaction), using a complementary primer for a designated microsatellite (Nilkanta et al., 2017). In the Stylosanthes genus, ISSR markers have been considered more efficient than both RAPD and STR markers to detect genetic variability (Nagaich and Chandra, 2009). Thus, our study aimed to evaluate the genetic diversity distribution in four populations of S. scabra in the semiarid region of Pernambuco state from Brazil using ISSR markers.
Material and Methods
The samplings were carried out in September 2015. Samples of 76 S. scabra plants were used from 4 natural populations in the semiarid region of the Pernambuco state, Brazil (Table 1) and 19 individuals represented each population.
| Populations | Latitude | Longitude | Populations characteristics |
|---|---|---|---|
| Santa Cruz do Capibaribe | 08º21'01” a 08º21'46” | 40º20'19” a 40º21'52” | Pasture |
| Floresta | 08º31’77” a 08º43’24” | 38º28’06” a 38º29’22” | Caatinga (road margin) |
| Sertânia | 08º00'52” a 08º32'35” | 36º27'58” a 37º36'28” | Caatinga (road margin) |
| Petrolina | 08º58'32” a 08º59'03” | 40º16'12” a 40º45’16” | Plants on the banks of the irrigation canal |
Table 1: Stylosanthes scabra populations studied in Pernambuco state, Brazil
For DNA extraction, young leaves were collected following the methodology adjusted for this genus by Santos-Garcia et al. (2012). The DNA extracted was quantified by comparison with lambda standards (Invitrogen, Carslbad, CA, USA) of concentrations known in ng μL-1 (100, 200 e 500) in 0.8% agarose gel. The purity integrity of the DNA samples was confirmed in a spectrophotometer under UV light (260/280 nm).
Seven primers of ISSR (Table 2) were used. The 25 μL reaction mixtures contained 1x PCR buffer (Invitrogen), 1.5 mM MgCl2, 0.8 μM of primer, 1 U of Platinum Taq DNA Polymerase (Invitrogen), 0.25 μM of each dNTP (Invitrogen), 25 ng of DNA template, and sterile distilled water to the total volume of 25μL. DNA amplifications were performed on thermocycler under the following conditions: 94°C for 4 min (initial denaturation), followed by 30 cycles of 94°C for 30s, 50°C for 1 min, and 72°C for 90s with a final extension step at 72°C for 7 min (Nagaich and Chandra 2009).
| Primer | Sequence | Loci | Polymorphism (%) |
|---|---|---|---|
| UBC 1 | ACACACAACACACACACT | 10 | 100 |
| UBC 2 | GAGSGSGAGAGAGAGAT | 12 | 100 |
| UBC 808 | AGAGAGAGAGAGAGAGC | 9 | 100 |
| UBC 810 | CTTCATTTCACTTCA | 16 | 100 |
| UBC 813 | GAGAGAGAGAGAGAGAA | 15 | 95.5 |
| UBC 879 | CTTCATTTCACTTCA | 12 | 100 |
| UBC 888 | BDBCACACACACACACA | 14 | 71,42 |
| Total | 88 | 95.27% |
Table 2: ISSR primers selected for Stylosanthes scabra populations, sequence, total fragment number per primer, polymorphism percentage (LP)
The amplification products were separated on a 2% agarose gel stained with Sybr Gold (Invitrogen), using the 100 bp marker (Invitrogen) and visualized under ultraviolet light and recorded on the digital photodocumentator Vilber Lourmat. The polymorphisms obtained using ISSR were tabulated according to the presence (1) or absence (0) of bands.
Genetic diversity was estimated using the Gen Alex 6.5 software (Peakall et al., 2012), with the analysis effective alleles number (Ne), Shannon Index (I), heterozygosity within populations (Hs), genetic differentiation coefficient (GST) and migrants number in the population (Nm).
Genetic identity and genetic distance among populations were computed using the model proposed by Nei (1978). Molecular variance analysis (AMOVA) was used to reveal the distribution of genetic diversity within and among populations. In this analysis, total genetic diversity was separated into two distinct hierarchical levels: difference among populations and among individuals within the population. The "Structure" program (Pritchard et al., 2000) based on Bayesian statistics was used to infer the groups number (k). The correlation between the geographic distance and genetic distance among S. scabra populations was made using the Genes program using 9999 permutations (Cruz, 2013).
Results and Discussion
The segregated amplicons numbers ranged from 9 for the UBC808 primer to 16 loci for UBC810 primer, which showed 16 loci (Figure 1) and reached 100% polymorphism (Table 2). The values found in our work are similar to other studies that used dominant markers in Mimosa caesalpinaefolia (Araújo et al., 2016), in Croton tetradenius (Almeida-Pereira et al., 2017) and in Desmanthus sp. (Costa et al., 2017).
The genetic differentiation coefficient was 0.334, considering all populations (Table 3). According to Lu et al. (2005), values of 0.30 represent a high degree of differentiation among populations, suggesting low rates of gene flow among populations. In contrast, there is no possibility of differentiating populations when the gene level flow is high (Ambiel et al., 2010). According to Collevatti et al. (2013), the genetic differentiation coefficient is one of several genetic parameters among populations, considering defining the species relation with the environment.
| Populations | Ne | I | HS | GST | NM |
|---|---|---|---|---|---|
| Santa Cruz do Capibaribe | 1.241 | 0.212 | 0.141 | ||
| Floresta | 1.235 | 0.206 | 0.138 | ||
| Sertânia | 1.150 | 0.134 | 0.089 | ||
| Petrolina | 1.242 | 0.197 | 0.136 | ||
| Média | 1.217 | 0.187 | 0.126 | ||
| Total | 1.434 | 0.445 | 0.334 | 0.500 |
Effective alleles number (Ne), Shannon index (I), Heterozygosity within populations (Hs), genetic differentiation coefficient (GST), migrants number in the population (Nm)
Table 3: Genetic differentiation parameters in four populations of Stylosanthes scabra in Pernambuco state, Brazil
The migrants number in the population was only 0.5 individuals per generation (Table 3), reaffirming the populations isolation. S. scabra is predominantly autogamous, so the low migrant’s number discards the occurrence of relevant genetic migration present in the process of population differentiation (Zamora et al., 2015). The genetic distance among populations varied from 0.244 between Sertânia and Santa Cruz do Capibaribe, and 0.505 between Santa Cruz do Capibaribe and Petrolina (Table 4). The closest populations were those with the lowest genetic distance, as well as the more distant populations geographically also presented greater genetic distance.
| Santa Cruz do Capibaribe | Floresta | Sertânia | Petrolina | |
|---|---|---|---|---|
| Santa Cruz do Capibaribe | 345 Km | 161 Km | 624 Km | |
| Floresta | 0.466 | 183 Km | 278 Km | |
| Sertânia | 0.244 | 0.275 | 464 Km | |
| Petrolina | 0.505 | 0.402 | 0.325 |
Table 4: Geographic distance and genetic distance for four populations of Stylosanthes scabra in Pernambuco state, Brazil.
The matrix correlation between genetic distance and geographic distance was significant according to Mantel test at 1% probability. However, the high correlation between both distances is uncommon in the literature (Jamnadas et al., 2006; Vigna et al., 2011). According to Zhao et al. (2012), environmental factors, mating system, population size and genetic flow are more important than geographic distance in population differentiation.
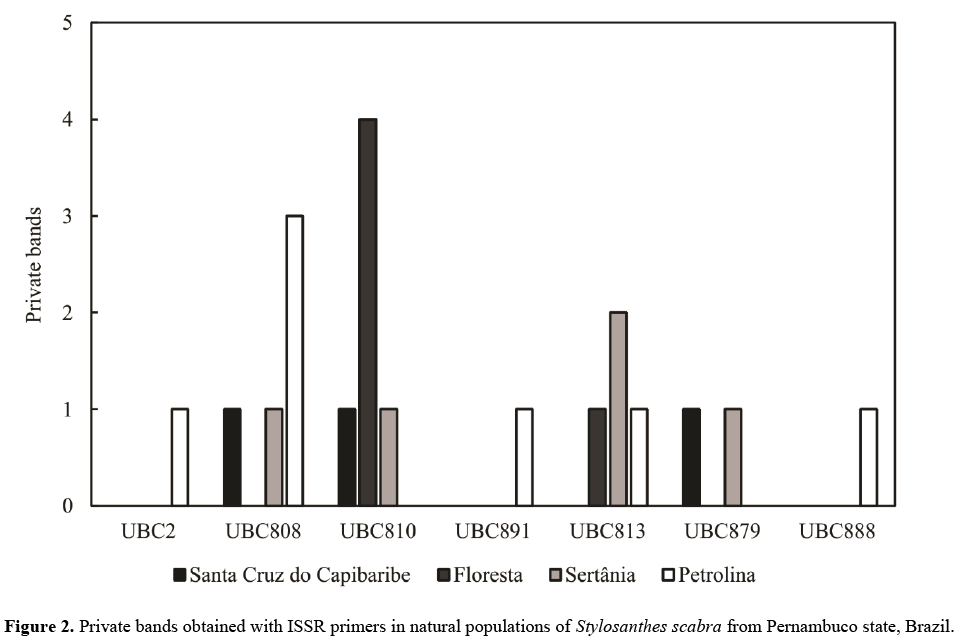
The Petrolina population presented the highest number of private bands (Figure 2) and greater variability, contributing to the appearance of DNA fragments (Szpiech and Rosenberg, 2011). According to Elstrand (2014), the occurrence of private bands is related to the average number of exchanged migrants per generations among populations and their presence (private bands) may indicate gene flow reduction.
The majority part of genetic variation (60%) was distributed among populations and 40% was distributed within populations, according to the AMOVA results. The species with autofecudation exhibit greater genetic diversity among the populations (Buzatti et al., 2012), whereas this is different for the plants with high rates of alogamy. In a study by Santos-Garcia et al. (2012) with microsatellite markers, S. capitata (which may present alogamia above 20%) did not present a correlation between the genetic distance and the collection sites, although few accesses by collection site were considered by these authors.
The minor genetic diversity within populations than among populations indicates the need for conservation and protection of all-natural populations in the evaluated region. Thus, it is unnecessary to collect many individuals by populations for S. scabra, but rather to collect few individuals in different populations, exploiting the maximum diversity in future collections.
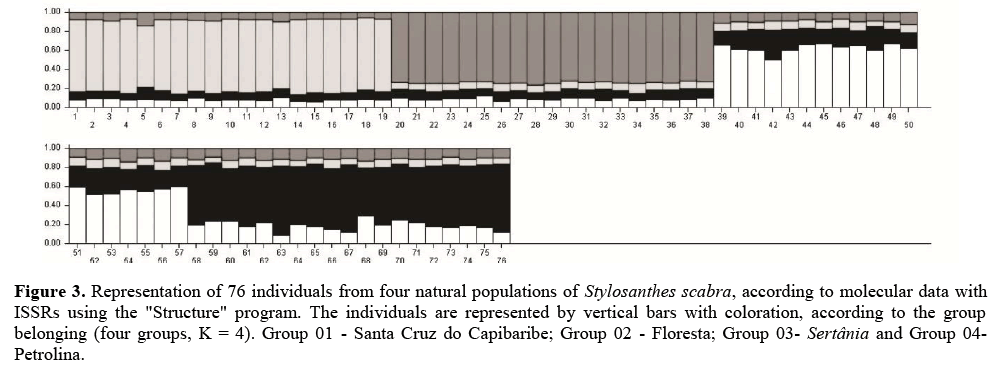
The population disposition can be visualized by the graphical distribution of species variability (Figure 3). Thus, four groups were also formed among the samples collected and followed the divisions by collection sites, differentiating the four populations. This grouping occurs by geographic isolation and due to autogamy in S. scabra (Chandra et al., 2009). In these cases, superior genotypes tend to stand out and populate the population with the largest number of offspring, increasing the similarity among them. Barros et al. (2005) also observed a trend of watershed separation for Stylosanthes accesses, collected in Bahia and Minas Gerais, both states from Brazil.
Figure 3: Representation of 76 individuals from four natural populations of Stylosanthes scabra, according to molecular data with ISSRs using the "Structure" program. The individuals are represented by vertical bars with coloration, according to the group belonging (four groups, K = 4). Group 01 - Santa Cruz do Capibaribe; Group 02 - Floresta; Group 03- Sertânia and Group 04- Petrolina.
Bayesian analyses showed a maximum K value = 4, confirming the well-defined genetic structure of the four populations. The results corroborate the expected, due to the low number of migrants and high genetic differentiation among the populations. According to Rossi et al. (2014), the genetic structuring of population occurs when the maximum K value coincides with the population’s number and the individuals of each population are grouped. Thus, there is intra-population genetic variability and the four sampling locations may be recognized as four distinct populations.
Finally, the ISSR markers used in our study were able to determine the genetic variability in S. scabra. There was a greater variability among populations than within populations, which showed the importance of S. scabra collections in different cities for the construction of germplasm banks. We suggest future collections, emphasizing Santa Cruz do Capibaribe and Petrolina (both municipalities in the state of Pernambuco, Brazil), to exploring the variability of this genus in these places.
Acknowledgment
We thank to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, Brazil)) for fellowship “Prodoutoral/CAPES” for the first author, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, Brazil), and Fundação de Amparo à Ciência e Tecnologia de Pernambuco (FACEPE, Pernambuco, Brazil) for research funding.
About the Authors
Corresponding Author
M.A. Lira Júnior
Universidade Federal Rural de Pernambuco, Departamento de Agronomia, Recife, PE, Brasil
- Email:
- mariolirajunior@gmail.com
References
- Almeida-Pereira CS, Silva AVC, Alves RP, Feitosa-Alcantara RB et al. (2017). Genetic diversity of native populations of Croton tetradenius Baill. using ISSR markers. Genet. Mol. Res. 16: 2-12. https://doi.org/10.4238/gmr16029602
- Ambiel AC, Machado Neto NB, Guaberto LM, Vanderlei TM (2010). Brachiaria germplasm dissimilarity as shown by RAPD markers. Crop Breed. Appl. Biot. 10: 55-64. https://doi.org/10.12702/1984-7033.v10n01a08
- Akinlade JA, Farinu GO, Agboola OO, Akingbade AA, et al. (2008). Nutritive value of four accessions of Stylosanthes scabra in the derived savanna zone of Nigeria. Trop. Grasslands 42: 120-123.
- Araújo FS, Pacheco MV, Vieira FA, Ferrari CS, et al. (2016). ISSR molecular markers for the study of the genetic diversity of Mimosa caesalpiniaefolia Benth. Idesia 34: 47-52.
- Barros AM, Faleiro FG, Karia CT, Shiratsuchi LS, et al. (2005). Variabilidade genética e ecológica de Stylosanthes macrocephala determinadas por RAPD e SIG. Pesq. Agropec. Bras. 40:899-909. https://doi.org/10.1590/s0100-204x2005000900010
- Brandão MB and Costa NMS (1979). O gênero Stylosanthes Swartz no Brasil. Epamig, Minas Gerais, 107p.
- Buzatti RSO, Ribeiro RA, Lemos Filho JP, and Lovato MB (2012). Fine-scale spatial genetic structure of Dalbergia nigra (Fabaceae), a threatened and endemic tree of the Brazilian Atlantic Forest. Genet. Mol. Biol. 35: 838-846. https://doi.org/10.1590/s1415-47572012005000066
- Calles T and Schultze-kraft R (2016). New species, nomenclatural changes and recent taxonomic studies in the genus Stylosanthes (Leguminosae): An update. Trop. Grasslands-Forrajes Tropicales 4: 122–128. https://doi.org/10.17138/tgft(4)122-128
- Chandra A (2009). Diversity among Stylosanthes species: Habitat, edaphic and agro-climatic affinities leading to cultivar development. J. Environ. Biol. 30: 471-478.
- Collevatti RG, Telles MPC, Nabout JC, Chaves LJ, et al. (2013). Demographic history and the low genetic diversity in Dipteryx alata (Fabaceae) from Brazilian Neotropical savannas. Heredity, 111: 97-105. https://doi.org/10.1038/hdy.2013.23
- Costa JC, Fracetto GGM, Fracetto FJC, Santos MVF, et al. (2017). Genetic diversity of Desmanthus sp accessions using ISSR markers and morphological traits. Genet. Mol. Res. 16: gmr16029667. https://doi.org/10.4238/gmr16029667
- Costa LC, Sartori ALB and Pott A (2008). Estudo taxonômico de Stylosanthes (Leguminosae - Papilionoideae - Dalbergieae) em Mato Grosso do Sul, Brasil. Rodriguésia 59: 547-572.
- Cruz CD (2013) GENES - a software package for analysis in experimental statistics and quantitative genetics. Acta Sci., Agron. 35: 271-276. http://dx.doi.org/10.4025/actasciagron.v35i3.21251
- Dias FTC, Bertini CHCM, Silva ANPM and Cavalcanti JJV (2015). Variabilidade genética de feijão-caupi de porte ereto e ciclo precoce analisada por marcadores RAPD e ISSR. Rev. Cienc. Agron. 46: 563-572. https://doi.org/10.5935/1806-6690.20150039
- Ellstrand NC (2014) Is gene flow the most important evolutionary force in plants? Am. J. Bot. 101: 737-753. https://doi.org/10.3732/ajb.1400024
- Fabrice CES, Soares Filho CV, Pinto MF, Perri SHV, et al. (2015). Recuperação de pastagens de Brachiaria decumbens degradada com introdução de Stylosanthes e adubação fosfatada. Rev. Bras. Saude Prod. Anim. 16: 758-771. https://doi.org/10.1590/s1519-99402015000400001
- Gonçalves AC, Reis CAF, Vieira FA and Carvalho D (2010). Estrutura genética espacial em populações naturais de Dimorphandra mollis (Fabaceae) na região norte de Minas Gerais. Rev. Bras. Bot. 33: 325-332. https://doi.org/10.1590/s0100-84042010000200013
- Gonzalez LM, Lopez RC, Fonseca I and Ramirez R (2000). Growth stomatal frequency, DM yield and accumulation of ions in nine species of grassland legumes grown under saline conditions. Pastos y Forrajes 23: 299- 308.
- Idowu OJ, Arigbede OM, Dele PA, Olanite JA, et al. (2013). Nutrients intake, performance and nitrogen balance of West African Dwarf sheep fed graded levels of toasted Enterolobium cyclocarpum seeds as supplement to Panicum maximum. Pak. J. Biol. Sci. 16: 1806–1810. https://doi.org/10.3923/pjbs.2013.1806.1810
- Jamnadass R, Mace ES, Hiernaux P, Muchugi A, et al. (2006) Population genetic responses of wild forage species to grazing along a rainfall gradient in the Sahel: A study combining phenotypic and molecular analyses. Euphytica 151: 431–445. https://doi.org/10.1007/s10681-006-9175-7
- Lu Y, Waller DM and David P (2005). Genetic variability is correlated with population size and reproduction in American wild-rice (Zizania palustris var. palustris, Poaceae) populations. Am. J. Bot. 92: 990-997. https://doi.org/10.3732/ajb.92.6.990
- Martuscello JA, Braz TGS, Silveira JM, Simeão RM, et al. (2015). Diversidade genética em acessos de Stylosanthes capitata. B. Indústr. Anim. 72: 284-289. https://doi.org/10.17523/bia.v72n4p284
- Mendonça ES, Lima PC, Guimarães GP, Moura WM, et al. (2017). Biological nitrogen fixation by legumes and N uptake by coffee plants. Rev. Bras. Cienc. Solo 41: e0160178. https://doi.org/10.1590/18069657rbcs20160178
- Mpanza TDE, Hassen A, Donkin EF and Nzuza WT (2014). Relative preference for, palatability and intake of Stylosanthes scabra accessions adapted in Pretoria. Trop. Grasslands-Forrajes Tropicales 2: 92–93. https://doi.org/10.17138/tgft(2)92-93
- Medrano M, López-Perea E and Herrera CM (2014). Population genetics methods applied to a species delimitation problem: endemic trumpet daffodils (Narcissus Section Pseudonarcissi) from the Southern Iberian Peninsula. Int. J. Plant Sci. 175: 501-517. https://doi.org/10.1086/675977
- Nagaich D and Chandra A (2009). Assessment of genetic diversity and identification of informative molecular markers for germplasm characterization in caribbean Stylo (Stylosanthes hamata). J. Plant Biochem. Biot. 18: 257–260. https://doi.org/10.1007/bf03263332
- Nagaich D, Tiwari KK, Srivastva N and Chandra A (2013). Assessment of genetic diversity and morpho-physiological traits related to drought tolerance in Stylosanthes scabra. Acta Physiol. Plant. 35: 3127-3136. https://doi.org/10.1007/s11738-013-1345-3
- Nei M (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583-590.
- Nilkanta H, Amom T, Tikendra L, Rahaman H, et al. (2017). ISSR marker based population genetic study of Melocanna baccifera (Roxb.) Kurz: a commercially important bamboo of manipur, north-east India. Scientifica 9: 1-9. https://doi.org/10.1155/2017/3757238
- Olafadehan AO, Adewumi MK and Andokunade AS (2014). Effects of feedingtannin-containing forage in varying proportion with concentrate on the voluntary intake, haematological and biochemical indices of goats. Trakia J. Sci. 12: 73–81.
- Oliveira RS, Queiróz MA, Romão RL, Silva GC, et al. (2016). Genetic diversity in accessions of Stylosanthes spp. using morphoagronomic descriptors. Rev. Caatinga 29: 101 – 112. https://doi.org/10.1590/1983-21252016v29n112rc
- Pangga IB, Chakraborty S and Yates D (2004). Canopy size and induced resistance in Stylosanthes scabra determine anthracnose severity at high CO2. Phytopathology 94: 221–227. https://doi.org/10.1094/phyto.2004.94.3.221
- Peakall R and Smouse PE (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research - an update. Bioinformatics 28: 2537-2539. https://doi.org/10.1093/bioinformatics/bts460
- Pen M, Savage DB, Nolan JV and Seng M (2013). Effect of Stylosanthes Guianensis supplementation on intake and nitrogen metabolism of Bos indicus cattle offered a basal diet of mixed rice straw and tropical grass. Anim. Prod. Sci. 53: 453–457. https://doi.org/10.1071/an11307
- Pritchard JK, Stephens M and Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945-959.
- Rossi FS, Rossi AAB, Dardengo JFE, Brauwers LR, et al. (2014). Diversidade genética em populações naturais de Mauritia flexuosa L. f. (Arecaceae) com uso de marcadores ISSR. Scientia florestales 42: 631-639.
- Santos-Garcia MO, Resende, RMS, Chiar L, Zucchi MI, et al. (2011). Mating systems intropical forage Stylosanthes capitata and Stylosanthes macrocephala (Aubl.) Sw. Euphytica 178: 185-193. https://doi.org/10.1007/s10681-010-0293-x
- Santos-Garcia MO, Toledo-Silva G, Sassaki RP, Ferreira TH, et al. (2012). Using genetic diversity information to establish core collections of Stylosanthes capitata and Stylosanthes macrocephala. Genet. Mol. Biol. 35: 847-861. https://doi.org/10.1590/s1415-47572012005000076
- Santana-Neto JA, Oliveira VS and Valença RL (2015). Leguminosas adaptadas como alternativa alimentar para ovinos no semiárido. Rev. Cienc. Agroveterinárias 14: 191-200.
- Szpiech ZA and Rosenberg NA (2011). On the size distribution of private microsatellite alleles. Theor. Popul. Biol. 80: 100-113. https://doi.org/10.1016/j.tpb.2011.03.006
- Srihari JM, Verma B, Kumar N, Chahota RK, et al. (2013). Analysis of molecular genetic diversity and population structure in sea buckthorn (Hippophae spp L.) from north-western Himalayan region of India. J. Med. Plants Res. 7: 3183-3196. https://doi.org/10.5897/JMPR12.1112
- Tufarelli V, Cazzato E, Ficco A and Laudadio V (2010). Evaluation of chemical composition and in vitro digestibility of appennine pasture plants using Yak (Bos grunniens) rumen fluid or faecal extract as inoculum source. Asian Austral. J. Anim. 23: 1587–1593. https://doi.org/10.5713/ajas.2010.10151
- Vieira FA, Sousa RF, Silva RAR, Fajardo CG, et al. (2015). Diversidade genética de Copernicia prunifera com o uso de marcadores moleculares ISSR. Agrária 10: 525-531. https://doi.org/10.5039/agraria.v10i4a5040
- Vigna BBZ, Jungmann L, Francisco PM, Zucchi MI, et al. (2011). Genetic diversity and population structure of the Brachiaria brizantha germplasm Trop. Plant Biol. 4: 157–169. https://doi.org/10.1007/s12042-011-9078-1
- Zamora-Tavares P, Vargas-Ponce O, Sánchez-Martínez J and Cabrera-Toledo D (2015). Diversity and genetic structure of the husk tomato (Physalis philadelphica Lam.) in Western Mexico. Genet. Resour. Crop Ev. 62: 141-153. https://doi.org/10.1007/s10722-014-0163-9
- Zhao X, Ma Y, Sun W, Wen X, et al. (2012) High genetic diversity and low differiantion of Michelia coriacea (Magnoliaceae), a critically endangered endemic in southeast Yunnan, China. Int. J. Mol. Sci. 13: 4396–4411. https://doi.org/10.3390/ijms13044396
Keywords:
Download:
Full PDF- Share This