Estimate of natural cross-pollination rate of Capsicum annuum using a codominant molecular marker associated with fruit pungency
Received: January 14, 2018
Accepted: February 24, 2018
Published: February 27, 2018
Genet.Mol.Res. 17(1): gmr16039887
DOI: 10.4238/gmr16039887
Abstract
The production of seeds with high genetic quality requires accurate estimates of the natural cross-pollination rate in cultivated Capsicum species. This is important in order to establish the most suitable isolation distance among fields of distinct genotypes, to minimize the risk of genetic contamination and to ensure that the agronomic and horticultural traits of an elite breeding material are preserved in a given seed lot. Cross-pollination studies in C. annuum are practically non-existent under Brazilian conditions. The objective of the present study was to estimate the natural rate of C. annuum cross-pollination in seed production fields established in the highland area of Central Brazil (“Cerrado”). A molecular marker (CA-Pun1), which allows the identification of cross-fertilization between C. annuum accessions with pungent and sweet fruits, was employed to identify hybrid seeds. This marker detects functional and non-functional alleles of an acyl-transferase (Pun1) gene, responsible to produce capsaicin (pungency) in placental tissue of C. annuum fruits. Seedlings of cultivar ‘Magali R’ (sweet fruits) were planted in the field at incremental distances from cultivar ‘BRS Garça’ (pungent fruits). Progenies obtained from ‘Magali R’ plants were genotyped with marker CA-Pun1. Approximately 10.8% of the fruits displayed at least one seed originated from cross-pollination. The overall natural cross-pollination rate in C. annuum was estimated at 1.2%, according to the percentage of hybrid seeds detected by molecular analysis. No significant differences were detected in the rate of cross-pollination among plants for distances of up to 7.2 m.
Introduction
The germplasm of the main domesticated pepper species of the genus Capsicum (C. annuum L., C. baccatum L., C. frutescens L. and C. chinense Jacq.) is characterized by a huge phenotypic variability, which is manifested in several traits of horticultural interest, including plant architecture, resistance to multiple diseases, size, aroma, shape, as well as biochemical composition of their fruits (Greenleaf, 1986; Boiteux et al., 1993; 1996; Pereira et al., 2011). This diversity represents an opportunity to direct use this germplasm in gene transfer programs via intra-specific and inter-specific crossings (Pickersgill, 1997).
Capsicum species have perfect (hermaphroditic) flowers, with male and female reproductive organs present in a single flower, which makes self-fertilization their major mating system (Pickersgill, 1997). The plants tend to self-pollinate prior to or during flower opening and the exposure of receptive stigmas to potential pollinators often occurs after self-pollination (Melo et al., 2014). However, different studies have shown that the cross-pollination rate in Capsicum is highly variable, ranging from 2 to 90% (Tanksley, 1984; Pickersgill, 1997). For example, in some cultivars of C. frutescens the possibility of cross-pollination is increased by morphological traits such as long styles, and it is reported to vary from 9 to 38% (Odland and Porter, 1941). Since the range of cross-pollination rate is wide, peppers are usually classified as having a facultative autogamous behavior.
The rate of natural cross-pollination is influenced by several factors, including genotype, geographical location, climatic conditions, predominant pollinators and spacing between plants (Tanksley, 1984). Hence, the isolation and/or the establishment of natural physical barriers between seed producing areas is very important for commercial seed production in order to minimize undesirable crossings, which may have negative impact on the genetic quality of the seeds. For the certified class of Capsicum seeds, the current Brazilian law requires an isolation of at least 400 meters between production fields. Even for hybrid seed production it is recommended to keep the physical separation of fields to avoid the risk of genetic contamination by insects that may penetrate the floral buds in search of pollen and/or nectar (Nascimento et al., 2006). This physical isolation procedure is recommended despite the fact that artificial pollination is carried out before the opening of the mature female flowers.
In Brazil, the information on cross-pollination rates naturally occurring in Capsicum species is practically non-existent. Cross-pollination rate estimates are important for evaluating the risk of genetic contamination and ensure that the agronomic and horticultural traits are preserved in the seed lot. Seed mixtures in commercial cultivars and hybrids may significantly alter their genetic purity, affecting resistance to biotic and abiotic stresses as well as yield, and other agronomic traits, causing potential negative impact on the profitability of growers (Nascimento et al., 2006).
Modern molecular marker techniques are allowing more precise estimates of cross-pollination, even when the genetic similarity of plant varieties involved in a mixture is high, or when seed mixtures include accessions with similar phenotypes (Sundaram et al., 2008). DNA techniques are, therefore, complementary tools to the conventional morphological descriptors often employed to estimate cross-pollination and evaluate genetic purity. In addition, molecular markers can be used to assess the genetic variability within and among species, with the potential to assist in the identification of new sources of variability in the development of new plant varieties (Ferreira and Grattaplagia, 1996). Molecular markers have been used in production systems to monitor the genetic quality of seed from cultivars and hybrids of different Capsicum species (Prince et al., 1995; Rodriguez et al., 1999; Ilbi 2003).
A peculiarity of a subset of Capsicum accessions is the presence of pungency, which is associated with the accumulation in the fruit placental tissue of at least six chemically related capsaicinoid (alkaloid) compounds (Wyatt et al., 2012). Capsaicin and its derivative dihydrocapsaicin are the two most potent pungency-related compounds (Supalkova et al., 2007). Their presence/absence and their relative contents in the fruit are traits of extreme interest for the pepper sauce and processing industry. Capsicum breeding programs have a dual interest in the development of both pungent chili pepper varieties (with high levels of capsaicin and dihydrocapsaicin) as well as sweet varieties (with no pungency at all) intended for the fresh and paprika market (Wyatt et al., 2012; Reddy et al., 2014).
Molecular markers linked to a gene encoding an acyl-transferase (named as Pun1 gene) that was found to be associated with the accumulation of capsaicin in Capsicum fruits have recently been developed by different research groups (Stewart et al., 2005; Ohse et al., 2010; Wyatt et al., 2012; Han et al., 2013; Reddy et al., 2014). Pun1 gene was identified as the single most important genetic factor related to pungency in Capsicum fruits (Han et al., 2013; Reddy et al., 2014). More recently, the precise chromosomal localization of this gene has been determined after obtaining the complete Capsicum genome sequence (Kim et al., 2014). Therefore, codominant molecular markers derived from this locus may be efficiently employed for the estimation of natural crossing rates and genetic purity of Capsicum seeds. In this context, the main purpose of this study was to estimate the rate of natural cross-pollination between accessions of C. annuum using a codominant molecular marker (CA-Pun1) associated with fruit pungency.
Material and Methods
Geographical location of the assay and environmental conditions
The present study was carried out between April and December 2012, at the experimental field and in the Seed Technology and Genomic Analysis Laboratories of the Embrapa Vegetable Crops in Brasília-DF, Brazil (Latitude 15° 56’’, Longitude 48° 08’’, and 997 meters above sea level). According to the Koppen classification, the climate of the region is a Caw type (with dry and mild winter, and warm and humid summer).
Capsicum accessions and seedling production
Cultivars ‘BRS Garça’ (with pungent fruits) and ‘Magali R’ (with sweet fruits) were selected to estimate the rate of natural cross-pollination in C. annuum. Seedlings were produced in expanded polystyrene (Styrofoam) trays with 200 cells, containing agricultural substrate Bioplant® consisting of pine bark, manure, sawdust, vermiculite, rice husk ash, gypsum (calcium sulfate), calcium carbonate, magnesium, Yoorin® thermophosphate, and micronutrients. Seeds of ‘BRS Garça’ were sown at weekly intervals (4/18/2012, 5/2/2012, and 5/17/2012), aiming to provide a maximum flowering overlap between the cultivars, whereas seeds of ‘Magali R’ were sowed on 05/05/2012. The trays were kept under greenhouse conditions.
Field trial to estimate natural crossing rates
The study was carried out in an open field area of 336 m2. The experimental area was prepared conventionally, and soil corrections were made according to chemical analyses. Fertilization and overall cultivation methods followed the recommendations for the crop. Although different experimental design models have been described for the determination of cross-pollination rate (e.g. Oliveira, 1963; Giordano et al., 1991), it was decided, in the present study, to plant the contrasting cultivars in parallel rows (Beri et al., 1985). Each plot was composed of a row of 20 plants spaced 1.0 m apart. ‘BRS Garça’ seedlings were transplanted to the two central rows 40 days after sowing. ‘Magali R’ seedlings were then transplanted to the six rows to the left and six rows to the right of ‘BRS Garça’, at increasing distances of 1.2; 2.4; 3.6; 4.8; 6.0, and 7.2 meters.
Phenological observations and survey of potential pollinator insects
Flower anthesis occurred in early July. During the flowering phase rainfall was nil and the average maximum and minimum temperatures were 27.8ºC and 15.9ºC, respectively. The monthly average relative humidity was 48%. Weekly field visits were made in order to survey the diversity of pollinators in the experimental area. The presence of the insects in the flowers of the accessions was annotated. Insect samples were collected and kept in 70% alcohol for entomological analysis.
Method used to sample seeds under field conditions
Cross-pollination estimates were based on the analysis of seeds of the first ripe fruit harvested in each plant of the ‘Magali R’ cultivar, totaling a sample of 240 fruits. Seeds of each fruit were extracted manually with the aid of a stylet under laboratory conditions. Seeds were then dried, packed and stored appropriately for carrying out the subsequent experiments.
DNA extraction and sample composition
Sixteen seeds obtained from one fruit of each of the 240 ‘Magali R’ plants (= 240 families) were sowed in expanded polystyrene (Styrofoam) trays with 200 cells, containing Bioplant® agricultural substrate, making a total of 3,840 seedlings. The trays were kept in greenhouse conditions. Leaf samples from each seedling were collected for DNA extraction when the seedlings displayed 3 to 4 well-expanded true leaves. DNA analyses were performed with two types of samples: (1) bulk extracted DNA samples (mixture of leaf tissue from 16 seedlings per family) to detect families with positive indication of crosses between ‘Magali R’ and ‘BRS Garça’; (2) individual analysis of 16 seeds of each family that displayed positive result for cross-pollination. Genomic DNA was extracted from young leaves using the CTAB (cetyltrimethylammonium bromide) method with some minor modifications (Boiteux et al., 1999). Samp(les of individual leaves (1 cm diameter leaf disc) or bulk (16 leaf discs per family) were conditioned in a microtube and macerated in an electronic shaker. To this macerate were added 750 μL of CTAB buffer [2% (w/v); 1.4 M NaCl; 100 mM Tris-HCl, 20 mM EDTA; 0.2% (v/v) β-mercaptoethanol] and then the mixture was homogenized using the apparatus Precellys®24. The pH was adjusted to 8.0 and the total volume was heated to 65°C in a water bath for ten minutes. A total of 750 μL of chlorophyll (chloroform/isoamyl alcohol 24:1 v/v) was added to the microtube, which was vortexed and centrifuged at 9,000 rpm for five minutes. In the following step, 600 μL of the supernatant were removed to a 1.5 mL microtube. The DNA in the aqueous phase was then precipitated by adding 300 μL of ice-cold isopropanol. The mixture was subjected to gentle stirring and subsequent centrifugation at 12,000 rpm for 13 minutes. The supernatant was discarded and the DNA at the bottom of the tube was washed with 70% ethanol aiming to remove the isopropanol and salt residues. The microtubes were placed for 20 minutes in an oven at 37°C. The DNA was dissolved in 300 μL of TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) and incubated for 24 hours at 4°C. The concentration and quality of the genomic DNA were estimated by optical density in NanoDrop® 1000 spectrophotometer (Thermo Scientific) according to the manufacturer's recommendations.
Polymerase chain reaction (PCR) assays
PCR reactions using as template genomic DNA were performed with marker CA-Pun1. This marker detects a major deletion on Pun1 gene (Ohse et al., 2012). C. annuum plants with the deletion (recessive defective allele) produce sweet fruits. CA-Pun1 is based on a triple primer system specific for the amplification of allelic variants of the Pun1 gene (Ohse et al., 2012), which encodes a presumably acyltransferase-like enzyme (GenBank accession number gb|EF104910), previously identified as being essential for the capsaicin production (Stewart et al., 2005). The set of primers was the following: ‘F1’: 5’-CGT-GAT-CAT-ACG-ACA-ACA-ACT-TTA-GT-3’; ‘F2’: 5’-TTT-AGG-TCA-TAC-CGC-TCC-CCG-AA-3’, and ‘R’: 5’-TGA-GGC-GTT-TAA-GAG-TAG-CTT-CTA-CAA-AT-3’ (Ohse et al., 2012). The sense primer ‘F1’ amplifies only the functional allele whereas the sense primer ‘F2’ amplifies only the defective allele. The antisense primer ‘R’ is common to both the defective allele and the functional allele of the presumed acyltransferase. The use of the system with three primers allows revealing a codominant polymorphism (a 300 bp amplicon present exclusively in plants with pungent fruits) and a defective allelic variant of 200 bp (present only in plants with non-pungent or sweet fruits) (Ohse et al., 2012). PCR assays were carried out in thermocyclers GeneAmp PCR System 9700 (Applied Biosystems) with the following program: 15 minutes at 95°C for denaturation, followed by 35 consecutive cycles of 30 seconds at 94°C, 3 minutes at 62°C, 2 minutes at 72°C, and a final step of 15 minutes at 68°C to ensure that any remaining single-strand DNA is fully extended. For each amplification reaction, the following concentration of each reagent was used: 2.0 μL of DNA at 90 ng/μL; 9.98 μL of autoclaved milli-Q water; 2.0 μL 10X Buffer with 15 mM MgCl2; 3.2 μl of 2.5 mM dNTP; 2.0 μL of 2.5 mM bovine serum albumin (BSA); 0.31 μL of each primer 3.2 mM and 0.2 μL of Taq DNA polymerase 5U/μL, with a final reaction volume 20.31 μL. Amplicons were separated on 1% agarose gel stained with ethidium bromide (5 μg/mL in water). The results were visualized in UV light and the gel images were captured using a digital system (Gel Doc, BioRad, USA).
Efficiency of marker CA-Pun1 for cross-pollination detection
For optimization of DNA analysis, the molecular marker CA-Pun1 was initially employed to evaluate the ability to detect at least one hybrid seed of the ‘Magali R’ × ‘BRS Garça’ cross in DNA blends of 16 seeds per family. For this, DNA samples of ‘BRS Garça’ e ‘Magali R’ were adjusted to the same concentration (40 ng/μL) and a synthetic DNA mixture of the two cultivars was prepared in the 1:15 ratio. Marker CA-Pun1 was then used to test the ability of identifying the ‘BRS Garça’ allele associated with pungency (Pun1 allele) in the synthetic samples, using as controls DNA samples extracted individually from: ‘BRS Garça’ (cultivar homozygous for the functional Pun1 allele – pungent fruits); ‘Magali R’ (cultivar homozygous for the defective Pun1 allele – sweet fruits); ‘CNPH 990’ and ‘CNPH 2725’ (inbred lines homozygous for the functional Pun1 allele – pungent fruits); ‘CNPH 196’ and ‘CNPH 581’, (inbred lines homozygous for the defective Pun1 allele – sweet fruits); ‘CNPH 305-1’ and ‘CNPH 653-1’ (inbred lines heterozygous at the Pun1 locus – pungent fruits). DNA extractions of these accessions were performed essentially as described above.
Experimental design and statistical analysis
The experimental design was a randomized complete block design, with six treatments and two replications. The estimate of cross-pollination rate was based on the genotyping of 16 seedlings of all ‘Magali R’ families which tested positive for the Pun1 pungency allele in the bulk analysis. The data were submitted to analysis of variance (ANOVA) and the means were compared by the Tukey’s test at 5% of probability, using the GENES® software package (Cruz, 2001).
Results
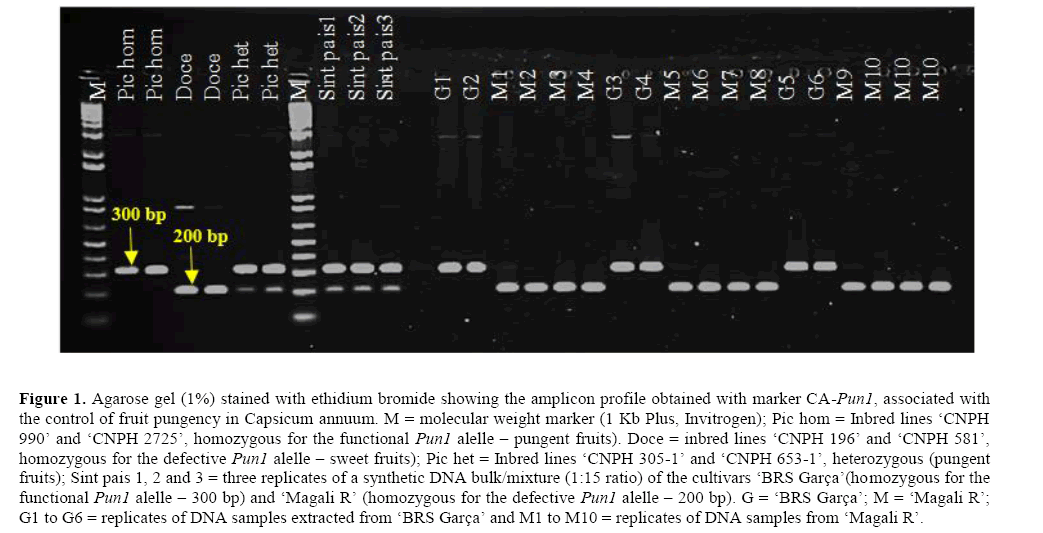
A molecular marker (CA-Pun1) able to detect functional and defective alleles of the Pun1 gene (which is associated with the genetic control of pungency) was used to estimate the natural cross-pollination rate in C. annuum (Ohse et al., 2012). Initially, the ability of this marker system to identify at least one hybrid seed in mixtures of 16 seeds per family was evaluated by DNA analysis of synthetic bulks formed by a mixture of genomic DNA extracted from the cultivars ‘Magali R’ and ‘BRS Garça’. The controls employed in this first set of experiments allowed the precise identification of both alternative alleles of the Pun1 gene (Figure 1). As expected, the control inbred lines with pungent fruits (‘CNPH 990’ and ‘CNPH 2725’) were found to be homozygous for the functional Pun1 allele (300 bp amplicons), whereas the control inbred lines with sweet fruits (‘CNPH 196’ and ‘CNPH 581’) were homozygous for the defective Pun1 allele (200 bp amplicons). Also, as expected, hybrid control accessions with pungent fruits (‘CNPH 305-1’ and ‘CNPH 653-1’) were heterozygous at the Pun1 locus (displaying both the 200 and 300 bp amplicons). The analyses of a synthetic DNA bulk/mixture (1:15 ratio) of the cultivars ‘BRS Garça’ and ‘Magali R’ allowed the confirmation of the high levels of efficiency of marker CA-Pun1 for detection of seeds of hybrid origin (Figure 1). These synthetic samples displayed amplicons corresponding to the two allelic variants of the Pun1 gene (Figure 1).
Figure 1: Agarose gel (1%) stained with ethidium bromide showing the amplicon profile obtained with marker CA-Pun1, associated with the control of fruit pungency in Capsicum annuum. M = molecular weight marker (1 Kb Plus, Invitrogen); Pic hom = Inbred lines ‘CNPH 990’ and ‘CNPH 2725’, homozygous for the functional Pun1 alelle – pungent fruits). Doce = inbred lines ‘CNPH 196’ and ‘CNPH 581’, homozygous for the defective Pun1 alelle – sweet fruits); Pic het = Inbred lines ‘CNPH 305-1’ and ‘CNPH 653-1’, heterozygous (pungent fruits); Sint pais 1, 2 and 3 = three replicates of a synthetic DNA bulk/mixture (1:15 ratio) of the cultivars ‘BRS Garça’(homozygous for the functional Pun1 alelle – 300 bp) and ‘Magali R’ (homozygous for the defective Pun1 alelle – 200 bp). G = ‘BRS Garça’; M = ‘Magali R’; G1 to G6 = replicates of DNA samples extracted from ‘BRS Garça’ and M1 to M10 = replicates of DNA samples from ‘Magali R’.
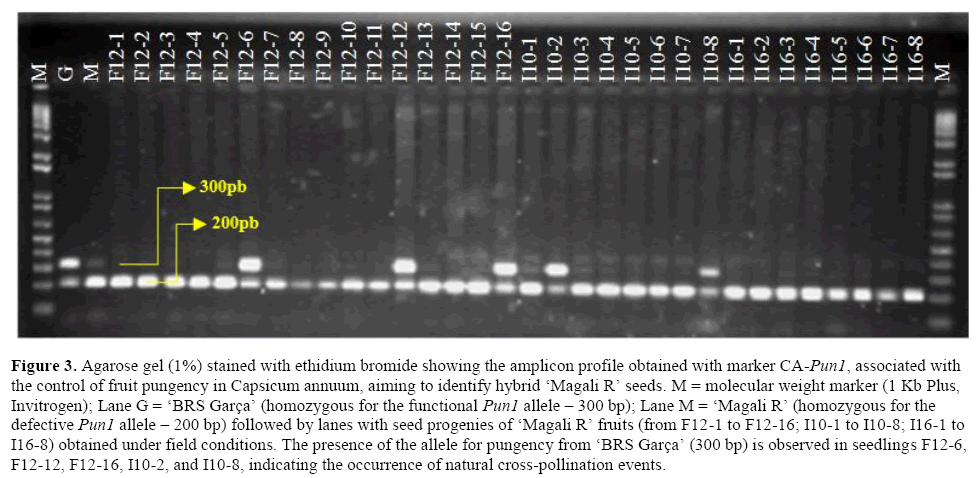
Based on this result, marker CA-Pun1 was used for genotyping DNA bulks extracted from 16 seedlings (= seeds) per family, obtained from a single fruit of ‘Magali R’ plants. The major objective of this assay was to identify the presence of families with at least one seed originated from the natural crossing between the two cultivars (‘Magali R’ and ‘BRS Garça’). Altogether, 240 families were genotyped for the presence of the functional Pun1 allele derived from the cultivar ‘BRS Garça’. This survey indicated that 26 families (10.8%) had at least one seed derived from a natural crossing between the two cultivars (Figures 2 and 3). Sixteen plants of each of the 26 bulks that tested positive for the pungency Pun1 allele were then individually genotyped with marker CA-Pun1. The observed marker genotypes were counted and an estimate of cross-pollination between ‘Magali R’ and ‘BRS Garça’ was computed according to the growing distances between plants of both cultivars (Table 1). The estimate of cross-pollination rate ranged from 0.94% to 1.56% between the two C. annuum accessions. A total of 46 seedlings originating from hybrid seeds were identified in 3,840 seedlings (= seeds) genotyped with CA-Pun1. This set of assays allowed us to estimate the average cross-pollination rate in C. annuum of 1.2% in Central Brazil. There was no significant difference between the crossings between ‘Magali R’ and ‘BRS Garça’ across all distances evaluated (between 1.2 and 7.2 m) (Table 1). The following orders of potential pollinating insects were observed visiting Capsicum flowers in our field surveys: Hymenoptera, Coleoptera, Lepidoptera, and Diptera.
| Experimental details and evaluations | Distance between plots of ‘BRSGarça’ and ‘Magali R’ | |||||
|---|---|---|---|---|---|---|
| 1.2m | 2.4m | 3.6m | 4.8m | 6m | 7.2m | |
| # of plants evaluated | 40 | 40 | 40 | 40 | 40 | 40 |
| # of seeds (progenies) genotyped with the molecular marker CA-Pun1 | 640 | 640 | 640 | 640 | 640 | 640 |
| # of ‘Magali R’-derived families positive for the ‘BRSGarça’ Pun1 allele (pungent fruit) | 4 | 6 | 5 | 5 | 3 | 3 |
| # of ‘Magali R’ heterozygous (hybrid) seeds for the Pun1 allele (sweet fruit) | 9 | 10 | 7 | 7 | 7 | 6 |
| % ‘Magali R’-derived families positive (= heterozygous) for the functional Pun1 allele (pungency) | 10.0% | 15.0% | 12.5% | 12.5% | 7.5% | 7.5% |
| % of natural cross-pollination between ‘BRSGarça’ and ‘Magali R’ | 1.41% a1 | 1.56% a | 1.09% a | 1.09% a | 1.09% a | 0.94% a |
Table 1: Estimates the natural cross-pollination rate between the Capsicum annuum cultivars ‘BRS Garça’ and ‘Magali R’ based on marker CA-Pun1.
Figure 2: Agarose gel (1%) stained with ethidium bromide showing the amplicon profile obtained with marker CA-Pun1, associated with the control of fruit pungency in Capsicum annuum. Bulks of DNA extracted from 16 seeds randomly collected from fruits of ‘Magali R’ (homozygous for the defective Pun1 allele – 200 bp). M = molecular weight marker (1 Kb Plus, Invitrogen); G = ‘BRS Garça’ (homozygous for the functional Pun1 allele – 300 bp); M = ‘Magali R’. Lanes A1 to A20; B1 to B20 and C1 to C6 = DNA bulks of families (16 seedlings) obtained from ‘Magali R’ plants which were naturally pollinated under field conditions.
Figure 3: Agarose gel (1%) stained with ethidium bromide showing the amplicon profile obtained with marker CA-Pun1, associated with the control of fruit pungency in Capsicum annuum, aiming to identify hybrid ‘Magali R’ seeds. M = molecular weight marker (1 Kb Plus, Invitrogen); Lane G = ‘BRS Garça’ (homozygous for the functional Pun1 allele – 300 bp); Lane M = ‘Magali R’ (homozygous for the defective Pun1 allele – 200 bp) followed by lanes with seed progenies of ‘Magali R’ fruits (from F12-1 to F12-16; I10-1 to I10-8; I16-1 to I16-8) obtained under field conditions. The presence of the allele for pungency from ‘BRS Garça’ (300 bp) is observed in seedlings F12-6, F12-12, F12-16, I10-2, and I10-8, indicating the occurrence of natural cross-pollination events.
Discussion
Knowledge of the natural cross-pollination rate is important to define the minimum physical distance among commercial seed production fields of Capsicum. Isolation is important to maintain the genetic purity of the seeds produced, which contributes to a higher quality of the commercialized seeds. So far, the estimates of the natural cross-pollination rates in C. annuum accessions are practically non-existent in Brazil.
Across all Brazilian regions, the commercial cultivation of peppers is still carried out, mainly, by small farmers in family-based agricultural systems. A common practice adopted by several farmers for the implementation of the pepper production fields within family-based agricultural systems has been the use of locally produced seeds obtained under field conditions. Often, these fields are planted without establishing a proper set of physical isolation strategies from the surrounding genotypes and farms. The consequence, the seeds used for planting have, in general, low physiological and genetic qualities, resulting in serious economic damage to the farmers in the following production cycles (Justino et al., 2015). In this context, the genetic quality of the seeds, determined by its phenotypic stability through crop generations, is of great agronomic importance. Farmers able to manage properly the seed production fields of a superior cultivar will maintain its most relevant horticultural traits throughout the generations, such as yield potential, resistance to diseases and pests, vegetative and reproductive cycles, and nutritional or sensorial attributes, such as the presence (or absence) of pungency.
Molecular markers have been widely used in genetic analyses for a wide range of purposes, such as identification of clones, cultivars, hybrids, levels of germplasm diversity, gene flow, and estimates of cross-pollination rates (Lacerda et al., 2002). Maker CA-Pun1 proved to be efficient in quantifying the natural cross-pollination rates of C. annuum (1.2%) under field conditions in the highland area of Central Brazil (“Cerrado”). In some C. frutescens varieties with more prominent stylus, the cross-pollination rates ranged from 9 to 38% (Odland and Porter, 1941). Other studies have shown that cross-pollination between Capsicum accessions may vary from 2 to 90% (Tanksley, 1984; Pickersgill, 1997). Differences in crossing rates in different Capsicum populations of the same species may be explained by variables such as planting time, spatial plant distribution, flowering overlap, population density of pollinating insects, genetic compatibility, among others. The abundant presence of a wide range of potential pollinator insects during the experiment is an aspect to be considered, especially in regard to the flight distance covered by the insects during the pollination process. The orders of potential pollinating insects observed in our field surveys were Hymenoptera, Coleoptera, Lepidoptera and Diptera. Many of them may be considered as potential cross-pollinator agents of Capsicum. However, a more detailed study should be carried out in order to clarify the relative importance of each group of insects as cross-pollinator agents.
Although the cross-pollination rate estimated in the present study in C. annuum (1.2%) may be considered low, it is likely that the current isolation requirement for the Capsicum seed production fields in Brazil is adequate in order to avoid genetic contamination through cross-pollination. It is likely that the minimum isolation distance required by Brazilian legislation (400 m) is more than enough to minimize the adverse effect of cross-pollination in seed production fields. Due to the potential for movement of the pollinator insect species found in this region, distances longer than those tested in the present experiment may be useful for a more accurate estimation of the adequate physical distance between Capsicum genotypes in certified seed fields. Exogenous contamination could be minimized by an abundant supply of desired pollen at the time the stigma is receptive. In this situation, the chance of foreign pollen be involved in the fertilization process is usually small. In practical terms, however, this strategy needs to be complemented by adequate field isolation (Peske et al, 2006), which reduces the risk of undesirable pollination.
Conclusion
The results obtained in the present study indicate that marker CA-Pun1 is useful for estimating the natural cross-pollination of C. annuum cultivars. This marker may also be applied for evaluating genetic purity and detect contaminant seeds of C. annuum, especially when there is potential for natural or undesirable crossing between cultivars of contrasting traits, such as sweet and pungent fruits.
About the Authors
Corresponding Author
W.M. Nascimento
National Center for Vegetable Crops Research (CNPH), Embrapa Vegetable Crops, CP 218, 70.275-970 BrasÃlia-DF, Brazil
- Email:
- warley.nascimento@embrapa.br
References
- Beri SM, Sohoo MS, Sharma HL (1985). Estimates of natural cross pollination in Egyptian clover. Euphytica 34: 147–151. Blum E, Liu K, Mazourek M, Yoo EY, et al. (2002). Molecular mapping of the C locus for presence of pungency in Capsicum. Genome 45: 702-705. https://doi.org/10.1139/g02-031 Boiteux LS, Nagata T, Dutra WP, Fonseca MEN (1993). Sources of resistance to tomato spotted wilt virus (TSWV) in cultivated and wild species of Capsicum. Euphytica 67: 89-94. https://doi.org/10.1007/bf00022729 Boiteux LS, Cupertino FP, Silva C, Dusi NA, et al. (1996). Resistance to potato virus Y (pathotype 1-2) in Capsicum annuum and Capsicum chinense is controlled by two independent major genes. Euphytica 87: 53-58. https://doi.org/10.1007/bf00022964
- Boiteux LS, Fonseca MEN, Simon PW (1999). Effects of plant tissue and DNA purification method on randomly amplified polymorphic DNA-based genetic fingerprinting analysis in carrot. J. Amer. Soc. Hort. Sci. 124: 32–38.
- Cruz CD (2001). Programa Genes: Versão Windows - Aplicativo Computacional em Genética e Estatística. Editora UFV, Viçosa.
- Ferreira ME, Grattapaglia D (1996). Introdução ao Uso de Marcadores Moleculares em Análise Genética. 2nd Ed. Brasília, DF: CENARGEN/EMBRAPA.
- Giordano LB, Marques MRC, and Melo PE (1991). Estimativa da taxa de cruzamento natural em ervilha. Hort. Bras. 9: 82-83.
- Han K, Jeong HJ, Sung J, Keum YS, et al. (2013). Biosynthesis of capsinoid is controlled by the Pun1 locus in pepper. Mol. Breed. 31: 537-548. https://doi.org/10.1007/s11032-012-9811-y
- Ilbi H (2003). RAPD markers assisted varietal identification and genetic purity test in pepper, Capsicum annuum. Sci Hort. 97: 211–218. https://doi.org/10.1016/s0304-4238(02)00158-9
- Justino EV, Boiteux LS, Fonseca MEN, Silva-Filho JG, et al. (2015). Determinação da maturidade fisiológica de sementes de pimenta dedo de moça Capsicum baccatum var. pendulum. Hort. Bras. 33: 324–333.
- Kim S, Park M, Yeom SI, Kim YM, et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46: 270 –278.
- Lacerda DR, Acedo MDP, Filho JPL, and Lovato NB (2002). A técnica de RAPD: Uma ferramenta molecular em estudos de conservação de plantas. Lundiana 3: 87–92.
- Melo AMT, Nascimento WM, Freitas RA (2014). Produção de Sementes de Pimenta. In: Nascimento WM (Ed.) Produção de sementes de hortaliças Vol. II. Embrapa, Brasília, DF, p. 169–197.
- Nascimento WM, Dias DCFS, and Freitas RA (2006). Produção de sementes de pimentas: Cultivo da pimenta. Inf Agrop. 27: 235.
- Odland ML, Porter AMA (1941). Study of natural crossing in pepper (Capsicum frutescens L.). J. Amer. Soc. Hort. Sci. 38: 585–588.
- Ohse BJG, Fuscaldi JL, Buso GSC, Carvalho SIC, et al. (2010). Ausência de ardor em pimenta causada por SNPs detectados no gene Pun1 de Capsicum annuum. In: 56º Anais Congresso Brasileiro de Genética, Guarujá, SP.
- Oliveira HA (1963). Ocorrência de alogamia em ervilha. Rev. Oler. 3: 83–90.
- Pickersgill B (1997). Genetic resources and breeding of Capsicum spp. Euphytica 96:129-133. https://doi.org/10.1007/978-3-319-06532-8_4
- Pereira MJZ, Massola-Junior NS, Sussel AAB, Sala FC, et al. (2011). Reação de acessos de Capsicum e de progênies de cruzamentos interespecíficos a isolados de Colletotrichum acutatum. Hort. Bras.29: 569–576.
- Prince JP, Lackney VK, Angeles C, Blauth J, et al. (1995). A survey of DNA polymorphism within the genus Capsicum and the fingerprinting of pepper cultivars. Genome 38: 224–231. https://doi.org/10.1139/g95-027
- Reddy UK, Almeida A, Abburi VL, Alaparthi SB, et al. (2014). Identification of gene-specific polymorphisms and association with capsaicin pathway metabolites in Capsicum annuum L. collections. PLoS One 9: e86393. https://doi.org/10.1371/journal.pone.0086393
- Rodriguez JM, Berke T, Engle L, Nienhuis J (1999). Variation among and within Capsicum species revealed by RAPD markers. Theor. Appl. Genet. 99: 147-156. https://doi.org/10.1038/sj.hdy.6881070
- Silva EF, Silva LM, Montalván R (2005). Crossing rate and distance in upland rice. Bragantia 64: 197-201. https://doi.org/10.1590/s0006-87052005000200005
- Stewart C, Kang B, Liu K, Mazourek M, et al. (2005). The Pun1 gene for pungency in pepper encodes a putative acyltransferase. Plant J. 42: 675–688. https://doi.org/10.1111/j.1365-313x.2005.02410.x
- Sundaram RM, Naveenkumar B, Biradar SK, Balachandran SM, et al. (2008). Identification of informative SSR markers capable of distinguishing hybrid rice parental lines and their utilization in seed purity assessment. Euphytica 163: 215-224. https://doi.org/10.15258/sst.2011.39.2.02
- Supalkova V, Stavelikova H, Krizkova S, Adam V, et al. (2007). Study of capsaicin content in various parts of pepper fruit by liquid chromatography with electrochemical detection. Acta Chemistry Slovenia 54: 55-59.
- Tanksley SD (1984). High rates of cross-pollination in chile pepper. HortScience 19: 580-582, 1984.
- Wang D and Bosland PW (2006). The genes of Capsicum. HortScience 41: 1169–1187.
- Wyatt LE, Eannetta NT, Stellari GM, Mazourek M (2012). Development and application of a suite of non-pungency markers for the Pun1 gene in pepper (Capsicum spp.). Mol. Breed.30: 1525-1529. https://doi.org/10.1007/s11032-012-9716-9
Keywords:
Download:
Full PDF- Share This