Decrypting the microRNA signatures in gastric cancer using high-throughput miRNA array coupled with systems biological approaches for precision medicine
Received: October 14, 2017
Accepted: November 08, 2017
Published: December 15, 2017
Genet.Mol.Res. 16(4): gmr16039845
DOI: 10.4238/gmr16039845
Abstract
MicroRNAs (miRNAs) are non-coding small RNA molecules that regulate the differential expression of genes. The expression of miRNAs is dysregulated in various types of cancers, including Gastric Cancer (GC), and has both prognostic and diagnostic potential. To interpret the role of miRNA expression in GC, we evaluated the expression profile using high-throughput miRNA array followed by in-depth systems biological analyses. Total RNA was isolated from 34 GC patients and 15 normal controls and hybridised and differentially expressed miRNAs in GC compared to normal controls were analysed. A total of 250 miRNAs were found to be differentially expressed in GC with a fold change ranging from + 2.5984 to – 3.5271 compared to normal gastric tissues. The differentially expressed miRNAs were further subjected to Ingenuity Pathway Analysis to understand the modulation of cellular and molecular functions and associated physiological system development and functions in GC. The Oncomirs such as miR-155-5p, miR-17-5p, miR-21-5p, miR-23a-3p, miR-24-3p, and miR-320b were significantly (P<0.05) upregulated and all the other miRNAs, mostly tumor suppressors, were significantly downregulated (P<0.05) leading to the reduction in cell death and the induction of gastric cancer, cellular growth and proliferation, cellular movement, cellular assembly and organization, DNA replication, recombination and repair, organismal injury and abnormalities and metastasis of tumor in GC compared to normal tissues. The high-throughput miRNA microarray coupled with systems biological analyses showed that miRNAs are dysregulated in GC, causing the initiation, progression, and metastasis of GC. In conclusion, the novel GC-inducing miRNA signatures decrypted in our study can further be exploited for precision medicine in clinics.
Introduction
Gastric cancer (GC) is the fifth most common and the third leading reason for cancer-related deaths around the globe (Suarez-Arriaga et al., 2017). The identification and development of novel methods using highly sensitive and specific biomarkers for rapid detection and monitoring the progression of GC could greatly reduce the mortality rate (Shekari et al., 2017). MicroRNAs (miRNAs/miRs) are non-coding small RNA molecules that regulate the differential expression of genes and RNA processing. The expression of miRNAs is tissue and tumor specific and dysregulated in cancers, including Gastric Cancer (GC), and plays an important role in tumorigenesis. miRNAs are evolutionarily conserved single-stranded RNA molecules (19-25 nucleotides) that play a vital role in post-transcriptional gene silencing by translational suppression or mRNA cleavage (Heyns and Kovalchuk, 2015). Till now, about 500 miRNAs have been recognised and ~3% of human genes encode for miRNAs that regulate 30% of protein coding genes. It was documented that a single miRNA could regulate about 200 different protein-coding genes and, hence, the effects of miRNAs are pleiotropic in nature (Ghorai and Ghosh, 2014). The abnormal or errant expression of miRNAs can affect the cellular and molecular homeostasis mechanisms necessary for the maintenance of normal physiological functions such as cellular growth, proliferation, differentiation, and cell death and potentially leading to various debilitating diseases or disorders (Manikandan et al., 2008; Pushparaj et al., 2008a). Studies have shown that miRNA expression positively correlates with variety of cancers and these non-coding transcripts function as both tumor suppressors and oncogenes (5-( Pushparaj et al., 2008b; Zhang et al., 2017)
The oncogenic miRNAs inhibit tumor suppressor miRs or genes that control cell differentiation or apoptosis and promote tumorigenesis are termed as Oncomirs (Palmero et al., 2011). On the other hand, some of the miRNA expression levels are considerably decreases during oncogenesis. These miRNAs are often precluding tumor development by negatively inhibiting oncogenes and/or genes that control cell proliferation and growth, cell differentiation or apoptosis. These anti-cancer miRNAs are called as tumor suppressors (Rong et al., 2017) and the reduction or deletion of a miRNA functioning as a tumor suppressor leads to tumor formation. The down regulation of mature tumor suppressor miRNAs can happen due to defective miRNA biogenesis and eventually leading to the increased translation of the oncoprotein. Hence, the profiling miRNA expression is pivotal due to their important regulatory roles in the cellular and molecular processes involved in cell death and survival, cell proliferation and growth, differentiation, development and apoptosis (Zhou et al., 2017). miRNA profiling by microarrays is a common high-throughput technique for the identification of biomarkers and drug targets. The data acquired from microarray analyses helps in the functional prediction of miRNAs by correlating miRNA expression patterns to corresponding gene and protein expression profiles. In the present study, microRNA profiling in GC was investigated using microarray followed by in-depth systems biological analyses in order to decrypt the differentially regulated oncomirs and tumor suppressors.
Material and Methods
Ethical statement
Written consent forms were obtained from Gastric cancer ppatients undergoing a surgical procedure. Medical ethical committee of KAU, Jeddah Saudi Arabia has approved this study (Reference#174-15).
Clinical samples and microarray
In this study, Formalin-fixed paraffin-embedded (FFPE) tissues biopsy samples from 34 gastric cancer patients were collected from King Abdulaziz university (KAU) hospital Jeddah. From healthy individuals 15 FFPE gastric biopsy samples were collected along with detailed clinical history from KAU hospital (Supplementary Table 1). Microarray analysis has been performed as described previously (Bibi et al., 2016) and data obtained in that study will be used in this study.
Microarray data analysis
Total RNA was isolated from 31 GC patients and 15 normal controls and hybridised using the manufacturer’s protocol on Genechip miRNA 4.0 (Thermofisher Scientific, USA). The differentially expressed miRNAs in GC compared to normal controls were analysed by Macrogen Inc (Seoul, Republic of Korea). Ingenuity Pathway (IPA) software (Qiagen, USA), was used to filter 140 analysis ready miRNAs out of 250 differentially expressed miRNAs in GC with a fold change ranging from + 2.5984 to – 3.5271 compared to the control. The 140 analysis ready miRNAs were subjected to IPA core analysis to deduce diseases and biological functions (Bio Functions) such diseases and disorders, molecular and cellular functions, physiological system development and function, pathological functions (Tox Functions), and unique miRNA networks.
The diseases and biological functions were further filtered using right tailed Fisher-Exact Test with a P value cut-off 0.05 with Benjamini-Hochberg (B-H) multiple testing corrections of the P values obtained for false discovery rates (FDR). Furthermore, the upregulated miRNAs and the down-regulated miRNAs were subjected to core analysis using IPA. Besides, comparison analysis module of IPA was utilized to compare the core analysis done using the upregulated miRNAs, down regulated miRNAs and the differentially expressed miRNAs (DEmiRs) to explore the regulation of diseases and bio functions, pathological and toxicological functions and the differential regulation of unique miRNA networks in gastric cancer. The differentially regulated diseases and bio-functions, pathological and toxicological functions were further clustered as heatmap and bar chart based on the activation Z-Score(12) provided by the IPA Knowledgebase, a powerful core technology based on Ingenuity Ontology.
Results and Discussion
miRNA expression profiles in gastric cancer and normal tissues samples
In the present study, we have uncovered the differential expression of miRNAs in GC compared to normal tissues using high-throughput miRNA arrays coupled with in-depth systems biological analyses. About 250 miRNAs were differentially regulated in GC compared to normal tissues. More importantly, the oncomirs such as miR-155-5p, miR-17-5p, miR-21-5p, miR-23a-3p, miR-24-3p, and miR-320b were significantly (P<0.05) upregulated in GC compared to normal tissues. Conversely, all the other miRNAs were significantly (P<0.05) downregulated in GC compared to normal tissues. The dysregulation of miRNA expression has been observed in various cancers including GC (Suarez-Arriaga et al., 2017; Shekari et al., 2017; Zhou et al., 2017). Approximately half of the annotated human miRNAs are found to be located in the genomic regions associated with cancer (Heyns and Kovalchuk, 2015). In normal tissues, appropriate miRNA transcription, processing and binding to target mRNAs to repress the oncogenic protein translation and subsequently maintain normal rates of cell death and survival, cellular growth and proliferation, differentiation and development (Palmero et al., 2011; Zhou et al., 2017). On the contrary, the oncogenesis is initiated by the increased expression of oncomirs and the down regulation of tumor suppressor miRNAs in cancer ( Zhang et al., 2017).
Regulatory networks
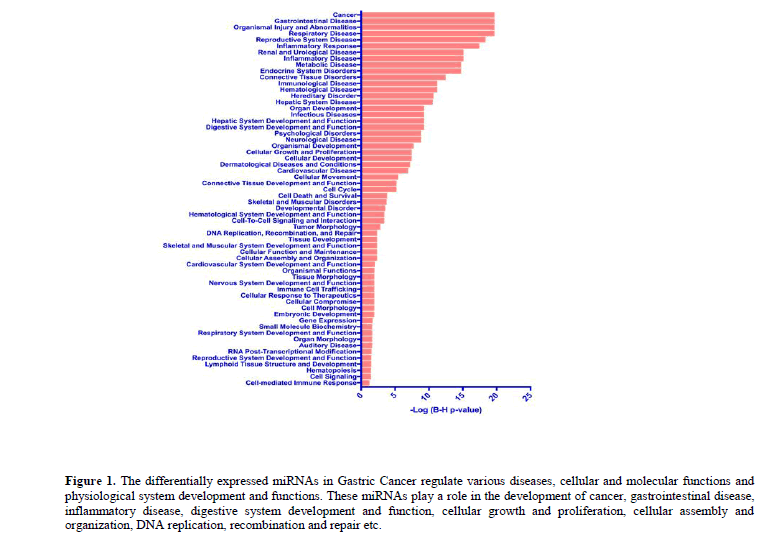
Systems biological analyses using the IPA revealed that these differentially regulated miRNAs in GC play a substantial role in the development of cancer, gastrointestinal disease, inflammatory disease and other debilitating diseases (Figure 1).
Figure 1: The differentially expressed miRNAs in Gastric Cancer regulate various diseases, cellular and molecular functions and physiological system development and functions. These miRNAs play a role in the development of cancer, gastrointestinal disease, inflammatory disease, digestive system development and function, cellular growth and proliferation, cellular assembly and organization, DNA replication, recombination and repair etc.
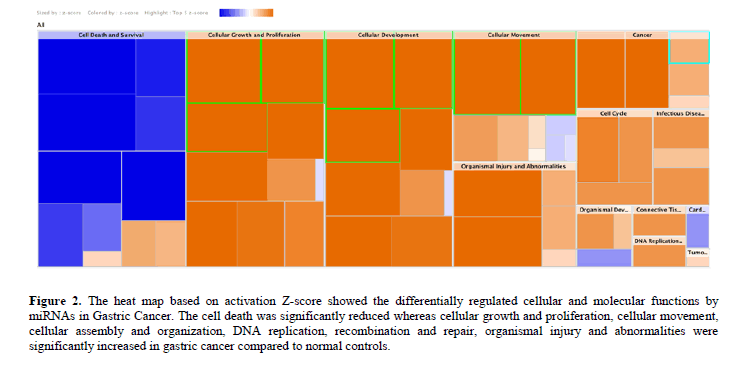
Besides, the cell death was significantly (P<0.05) reduced whereas cellular growth and proliferation, cellular movement, cellular assembly and organization, DNA replication, recombination and repair, organismal injury and abnormalities were significantly increased in GC compared to normal tissues (Figure 2).
Figure 2: The heat map based on activation Z-score showed the differentially regulated cellular and molecular functions by miRNAs in Gastric Cancer. The cell death was significantly reduced whereas cellular growth and proliferation, cellular movement, cellular assembly and organization, DNA replication, recombination and repair, organismal injury and abnormalities were significantly increased in gastric cancer compared to normal controls.
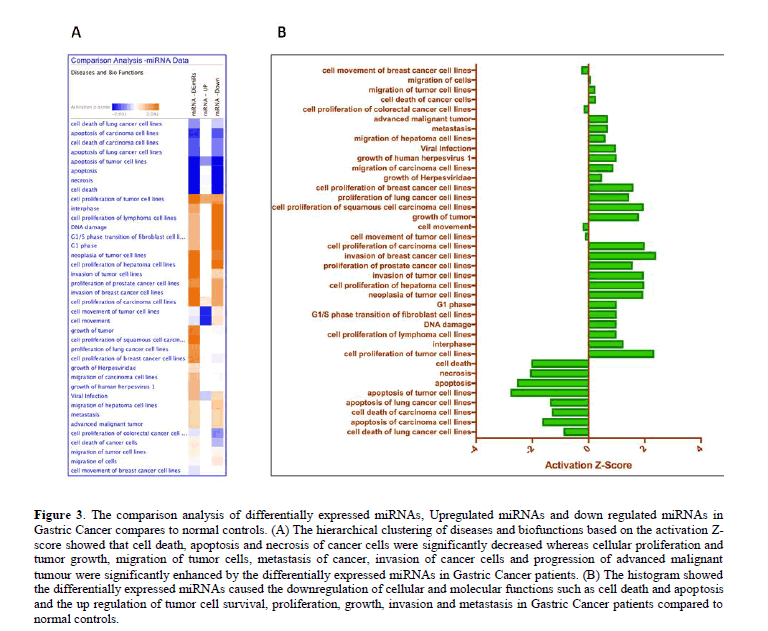
The hierarchical clustering of diseases and biofunctions based on the activation Z-score derived from the comparison analysis module in IPA showed that cell death, apoptosis and necrosis of cancer cells were significantly decreased (P<0.05) whereas the cellular proliferation and tumor growth, migration of tumor cells, metastasis of cancer, invasion of cancer cells and progression of advanced malignant tumour were significantly enhanced (P<0.05) by the differentially expressed miRNAs in GC patients. The differentially expressed miRNAs caused the downregulation of cellular and molecular functions such as cell death and apoptosis and the up regulation of tumor cell survival, proliferation, growth, invasion and metastasis in GC patients compared to normal controls (Figure 3).
Figure 3: The comparison analysis of differentially expressed miRNAs, Upregulated miRNAs and down regulated miRNAs in Gastric Cancer compares to normal controls. (A) The hierarchical clustering of diseases and biofunctions based on the activation Z-score showed that cell death, apoptosis and necrosis of cancer cells were significantly decreased whereas cellular proliferation and tumor growth, migration of tumor cells, metastasis of cancer, invasion of cancer cells and progression of advanced malignant tumour were significantly enhanced by the differentially expressed miRNAs in Gastric Cancer patients. (B) The histogram showed the differentially expressed miRNAs caused the downregulation of cellular and molecular functions such as cell death and apoptosis and the up regulation of tumor cell survival, proliferation, growth, invasion and metastasis in Gastric Cancer patients compared to normal controls.
The miRNAs in GC involved in the differential regulation of various diseases and biofunctions were listed in the supplementary table.
Gene expression
In our study, the increased expression of oncomiRs such as miR-155-5p, miR-17-5p, miR-21-5p, miR-23a-3p, miR-24-3p, and miR-320b and the down regulation of most of the tumor suppressors such as let-7a-5p, miR-16-5p, miR-29b-3p, miR-125b-5p, miR-130a-3p, miR-143-3p, miR-145-5p, miR-185-5p, miR-378a-3p, miR-638 and other functionally related miRNAs could mostly explain the mechanism behind the initiation, progression and metastasis of GC.
Studies have shown that miR-155-5p overexpression is associated with variety of cancers including GC (Feng, 2016). Both miR-155 and miR-21-5p were upregulated and promote tumor whereas the miR-145-5p was down regulated in GC (Liu et al., 2010). miR-23a-5p was upregulated in various cancers including GC and stimulates cancer growth (Zhu et al., 2010). Phosphatase and Tensin Homolog (PTEN), a tumor suppressor gene, induces apoptosis through the P13K-Akt-PKB pathway and it is inhibited by the over expression of oncogenic miR-17-5p (Zhou et al., 2011). Over expression of mir-17-5p down regulates PTEN and leads to tumor development. The over expression of mir-24-3p was shown to attenuate the levels of pro-apoptotic factors such as Cas 9 and APAF1, leading to cell proliferation, growth and tumor progression (Navarro and Lieberman, 2015; Xie et al., 2013).
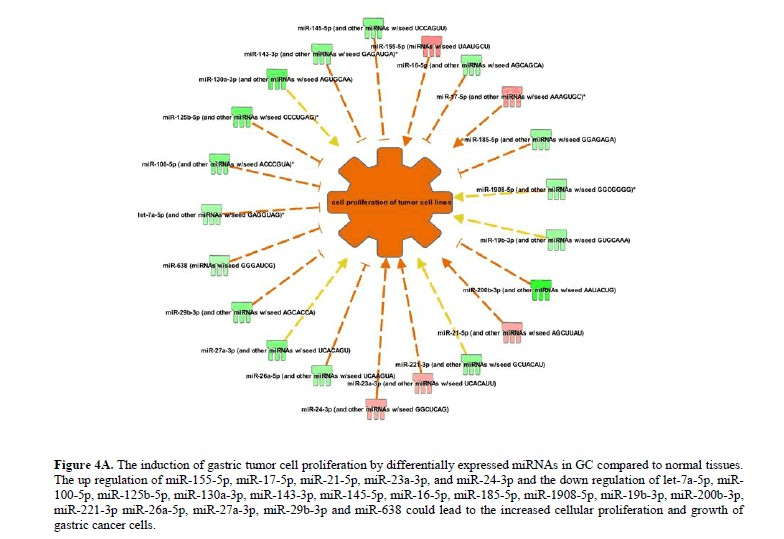
Interestingly, the miR-320a is a tumor suppressor whereas its homologue, miR-320b promotes tumor progression and invasion in different types of cancers (Lv et al., 2017; Zhou et al., 2015). The miRNA network regulating the cellular proliferation and growth of tumor cells was significantly upregulated (P<0.05) in GC. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p, miR-23a-3p, and miR-24-3p and the down regulation of let-7a-5p, miR-100-5p, miR-125b-5p, miR-130a-3p, miR-143-3p, miR-145-5p, miR-16-5p, miR-185-5p, miR-1908-5p, miR-19b-3p, miR-200b-3p, miR-221-3p miR-26a-5p, miR-27a-3p, miR-29b-3p and miR-638 leads to the increased cellular proliferation and growth of gastric cancer cells (Figure 4A).
Figure 4A: The induction of gastric tumor cell proliferation by differentially expressed miRNAs in GC compared to normal tissues. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p, miR-23a-3p, and miR-24-3p and the down regulation of let-7a-5p, miR-100-5p, miR-125b-5p, miR-130a-3p, miR-143-3p, miR-145-5p, miR-16-5p, miR-185-5p, miR-1908-5p, miR-19b-3p, miR-200b-3p, miR-221-3p miR-26a-5p, miR-27a-3p, miR-29b-3p and miR-638 could lead to the increased cellular proliferation and growth of gastric cancer cells.
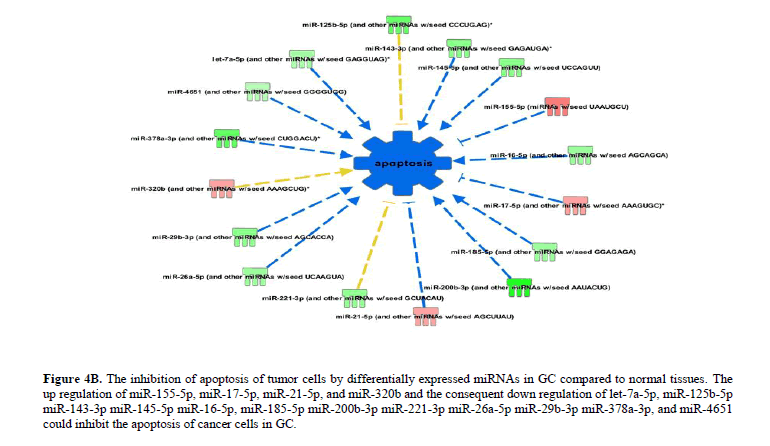
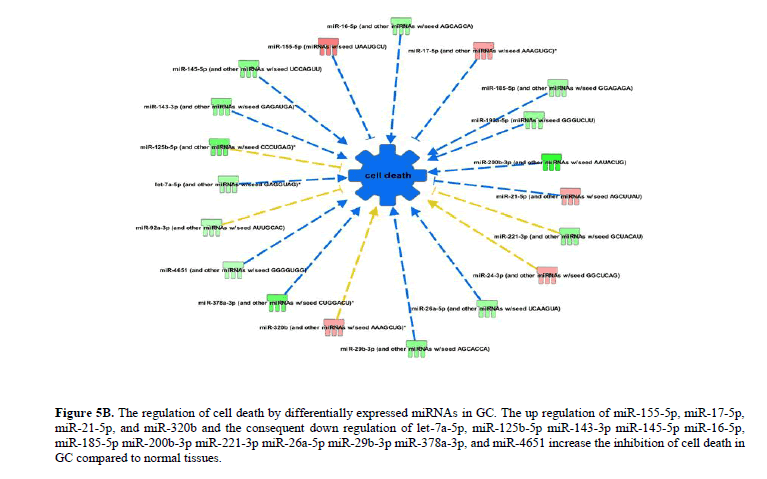
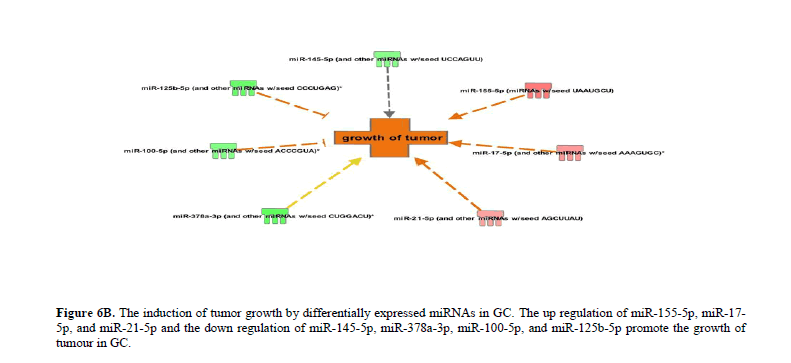
The differentially expressed miRNAs in GC significantly (P<0.05) attenuate apoptosis and promote cellular proliferation and growth. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p, and miR-320b and the consequent down regulation of let-7a-5p, miR-125b-5p miR-143-3p miR-145-5p, miR-16-5p, miR-185-5p miR-200b-3p miR-221-3p miR-26a-5p miR-29b-3p, miR-378a-3p, and miR-4651 increase the inhibition of apoptosis (Figure 4B), necrosis (Figure 5A) and cell death (Figure 5B) in GC. The up regulation of miR-155-5p, miR-17-5p, and miR-21-5p and the down regulation of miR-145-5p, miR-378a-3p, miR-100-5p, and miR-125b-5p promote the growth of tumour in GC compared to normal tissues (Figure 6A).
Figure 4B: The inhibition of apoptosis of tumor cells by differentially expressed miRNAs in GC compared to normal tissues. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p, and miR-320b and the consequent down regulation of let-7a-5p, miR-125b-5p miR-143-3p miR-145-5p miR-16-5p, miR-185-5p miR-200b-3p miR-221-3p miR-26a-5p miR-29b-3p miR-378a-3p, and miR-4651 could inhibit the apoptosis of cancer cells in GC.
Figure 5A: The inhibition of necrosis of cancer cells by differentially expressed miRNAs in GC. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p, and miR-320b and the consequent down regulation of let-7a-5p, miR-125b-5p, miR-143-3p, miR-145-5p, miR-16-5p, miR-185-5p, miR-200b-3p, miR-221-3p, miR-193a-5p, miR-26a-5p, miR-29b-3p, miR-378a-3p, and miR-4651 inhibit necrosis in GC compared to normal tissues.
Figure 5B: The regulation of cell death by differentially expressed miRNAs in GC. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p, and miR-320b and the consequent down regulation of let-7a-5p, miR-125b-5p miR-143-3p miR-145-5p miR-16-5p, miR-185-5p miR-200b-3p miR-221-3p miR-26a-5p miR-29b-3p miR-378a-3p, and miR-4651 increase the inhibition of cell death in GC compared to normal tissues.
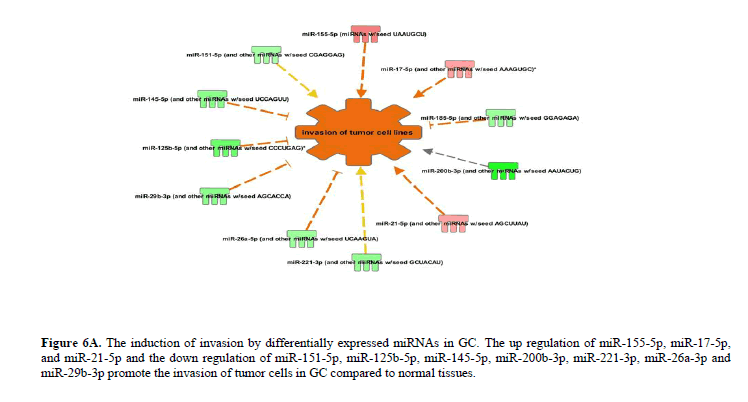
Figure 6A: The induction of invasion by differentially expressed miRNAs in GC. The up regulation of miR-155-5p, miR-17-5p, and miR-21-5p and the down regulation of miR-151-5p, miR-125b-5p, miR-145-5p, miR-200b-3p, miR-221-3p, miR-26a-3p and miR-29b-3p promote the invasion of tumor cells in GC compared to normal tissues.
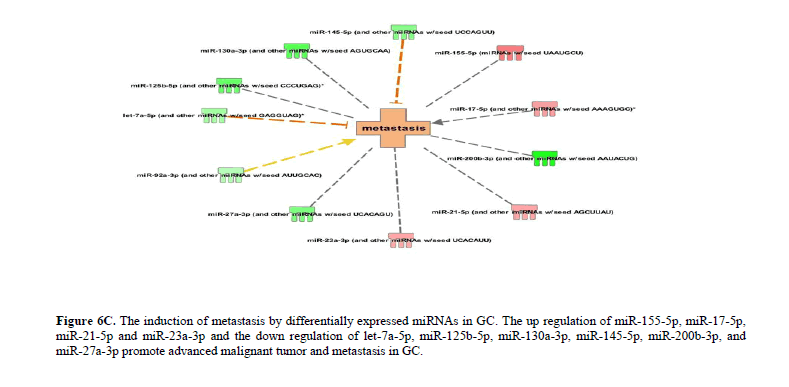
The up regulation of miR-155-5p, miR-17-5p, and miR-21-5p and the down regulation of miR-151-5p, miR-125b-5p, miR-145-5p, miR-200b-3p, miR-221-3p, miR-26a-3p and miR-29b-3p promote the invasion of tumor cells in GC (Figure 6B). Likewise, the up regulation of miR-155-5p, miR-17-5p, miR-21-5p and miR-23a-3p and the down regulation of let-7a-5p, miR-125b-5p, miR-130a-3p, miR-145-5p, miR-200b-3p, and miR-27a-3p promote advanced malignant tumor and subsequently leading to metastasis in GC (Figure 6C). The complete list of differentially expressed miRNAs regulating various cellular and molecular functions in GC has been provided in the Table 1 and Figure 7.
Figure 6C: The induction of metastasis by differentially expressed miRNAs in GC. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p and miR-23a-3p and the down regulation of let-7a-5p, miR-125b-5p, miR-130a-3p, miR-145-5p, miR-200b-3p, and miR-27a-3p promote advanced malignant tumor and metastasis in GC.
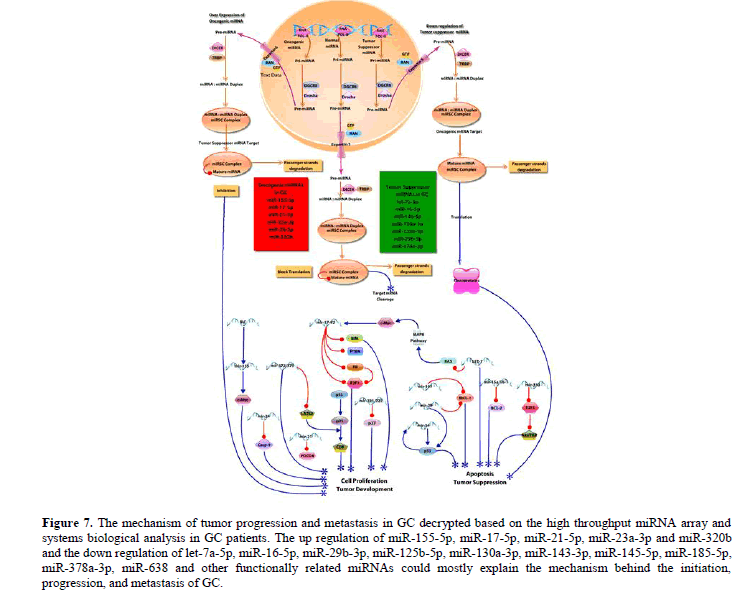
Figure 7: The mechanism of tumor progression and metastasis in GC decrypted based on the high throughput miRNA array and systems biological analysis in GC patients. The up regulation of miR-155-5p, miR-17-5p, miR-21-5p, miR-23a-3p and miR-320b and the down regulation of let-7a-5p, miR-16-5p, miR-29b-3p, miR-125b-5p, miR-130a-3p, miR-143-3p, miR-145-5p, miR-185-5p, miR-378a-3p, miR-638 and other functionally related miRNAs could mostly explain the mechanism behind the initiation, progression, and metastasis of GC.
| S.No | Diseases or Functions Annotation | P-Value | PredictedActivationState | Activz-score | Molecules | No ofMolecules |
|---|---|---|---|---|---|---|
| 1 | Apoptosis of tumor cell lines | 0.000227 | Decreased | -2.778 | let-7a-5p (and other miRNAs w/seedGAGGUAG),miR-143-3p (and other miRNAsw/seed GAGAUGA),miR-145-5p (and othermiRNAs w/seed UCCAGUU),miR-155-5p(miRNAs w/seed UAAUGCU),miR-16-5p(and other miRNAs w/seed AGCAGCA),miR-17-5p (and other miRNAs w/seedAAAGUGC),miR-185-5p (and other miRNAsw/seed GGAGAGA),miR-200b-3p (and othermiRNAs w/seed AAUACUG),miR-21-5p (andother miRNAs w/seed AGCUUAU),miR-221-3p (and other miRNAs w/seedGCUACAU),miR-29b-3p (and other miRNAsw/seed AGCACCA),miR-4651 (and othermiRNAs w/seed GGGGUGG) | 12 |
| 2 | Apoptosis | 0.00229 | Decreased | -2.555 | let-7a-5p (and other miRNAs w/seedGAGGUAG),miR-125b-5p (and othermiRNAs w/seed CCCUGAG),miR-143-3p(and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seedUCCAGUU),miR-155-5p (miRNAs w/seedUAAUGCU),miR-16-5p (and other miRNAsw/seed AGCAGCA),miR-17-5p (and othermiRNAs w/seed AAAGUGC),miR-185-5p(and other miRNAs w/seed GGAGAGA),miR-200b-3p (and other miRNAs w/seedAAUACUG),miR-21-5p (and other miRNAsw/seed AGCUUAU),miR-221-3p (and othermiRNAs w/seed GCUACAU),miR-26a-5p(and other miRNAs w/seed UCAAGUA),miR-29b-3p (and other miRNAs w/seedAGCACCA),miR-320b (and other miRNAsw/seed AAAGCUG),miR-378a-3p (and othermiRNAs w/seed CUGGACU),miR-4651 (andother miRNAs w/seed GGGGUGG) | 16 |
| 3 | Necrosis | 0.00037 | Decreased | -2.078 | let-7a-5p (and other miRNAs w/seedGAGGUAG),miR-125b-5p (and othermiRNAs w/seed CCCUGAG),miR-143-3p(and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seedUCCAGUU),miR-155-5p (miRNAs w/seedUAAUGCU),miR-16-5p (and other miRNAsw/seed AGCAGCA),miR-17-5p (and othermiRNAs w/seed AAAGUGC),miR-185-5p(and other miRNAs w/seed GGAGAGA),miR-193a-5p (miRNAs w/seed GGGUCUU),miR-200b-3p (and other miRNAs w/seedAAUACUG),miR-21-5p (and other miRNAsw/seed AGCUUAU),miR-221-3p (and othermiRNAs w/seed GCUACAU),miR-24-3p (andother miRNAs w/seed GGCUCAG),miR-26a-5p (and other miRNAs w/seedUCAAGUA),miR-29b-3p (and other miRNAsw/seed AGCACCA),miR-320b (and othermiRNAs w/seed AAAGCUG),miR-378a-3p(and other miRNAs w/seed CUGGACU),miR-4651 (and other miRNAs w/seed GGGGUGG) | 18 |
| 4 | Cell death | 0.00233 | Decreased | -2.04 | let-7a-5p (and other miRNAs w/seedGAGGUAG),miR-125b-5p (and othermiRNAs w/seed CCCUGAG),miR-143-3p(and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seedUCCAGUU),miR-155-5p (miRNAs w/seedUAAUGCU),miR-16-5p (and other miRNAsw/seed AGCAGCA),miR-17-5p (and othermiRNAs w/seed AAAGUGC),miR-185-5p(and other miRNAs w/seed GGAGAGA),miR-193a-5p (miRNAs w/seed GGGUCUU),miR-200b-3p (and other miRNAs w/seedAAUACUG),miR-21-5p (and other miRNAsw/seed AGCUUAU),miR-221-3p (and othermiRNAs w/seed GCUACAU),miR-24-3p (andother miRNAs w/seed GGCUCAG),miR-26a-5p (and other miRNAs w/seedUCAAGUA),miR-29b-3p (and other miRNAsw/seed AGCACCA),miR-320b (and othermiRNAs w/seed AAAGCUG),miR-378a-3p(and other miRNAs w/seed CUGGACU),miR-4651 (and other miRNAs w/seedGGGGUGG),miR-92a-3p (and other miRNAsw/seed AUUGCAC) | 19 |
| 5 | Apoptosis of carcinoma cell lines | 0.0000701 | Decreased | -1.649 | let-7a-5p (and other miRNAs w/seed GAGGUAG),miR-143-3p (and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-21-5p (and other miRNAs w/seed AGCUUAU),miR-221-3p (and other miRNAs w/seed GCUACAU),miR-4651 (and other miRNAs w/seed GGGGUGG) | |
| 6 | Apoptosis of lung cancer cell lines | 0.000299 | Decreased | -1.382 | miR-143-3p (and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-21-5p (and other miRNAs w/seed AGCUUAU),miR-221-3p (and other miRNAs w/seed GCUACAU),miR- 4651 (and other miRNAs w/seed GGGGUGG) | 5 |
| 7 | Cell death of carcinoma cell lines | 0.0000186 | Decreased | -1.31 | let-7a-5p (and other miRNAs w/seed GAGGUAG),miR-143-3p (and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-200b-3p (and other miRNAs w/seed AAUACUG),miR- 21-5p (and other miRNAs w/seed AGCUUAU),miR-221-3p (and other miRNAs w/seed GCUACAU),miR-4651 (and other miRNAs w/seed GGGGUGG) | 7 |
| 8 | Cell death of lung cancer cell lines | 0.0000667 | Decreased | -0.886 | miR-143-3p (and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-200b-3p (and other miRNAs w/seed AAUACUG),miR-21-5p (and other miRNAs w/seed AGCUUAU),miR-221- 3p (and other miRNAs w/seed GCUACAU),miR-4651 (and other miRNAs w/seed GGGGUGG) | 6 |
| 9 | Growth of tumor | 0.00269 | Increased | 1.791 | miR-100-5p (and other miRNAs w/seed ACCCGUA),miR-125b-5p (and other miRNAs w/seed CCCUGAG),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR- 155-5p (miRNAs w/seed UAAUGCU),miR- 17-5p (and other miRNAs w/seed AAAGUGC),miR-21-5p (and other miRNAs w/seed AGCUUAU),miR-378a-3p (and other miRNAs w/seed CUGGACU) | 7 |
| 10 | Neoplastic of tumor cell lines | 0.0021 | Increased | 1.949 | let-7a-5p (and other miRNAs w/seed GAGGUAG),miR-125b-5p (and other miRNAs w/seed CCCUGAG),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR- 17-5p (and other miRNAs w/seed AAAGUGC),miR-21-5p (and other miRNAs w/seed AGCUUAU) | 5 |
| 11 | Cell proliferation of squamous cell carcinoma cell lines | 0.0000955 | Increased | 1.963 | miR-100-5p (and other miRNAs w/seed ACCCGUA),miR-143-3p (and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-24-3p (and other miRNAs w/seed GGCUCAG) | 4 |
| 12 | Invasion of tumor cell lines | 0.00000348 | Increased | 1.968 | miR-125b-5p (and other miRNAs w/seed CCCUGAG),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-151-5p (and other miRNAs w/seed CGAGGAG),miR-155-5p (miRNAs w/seed UAAUGCU),miR-17-5p (and other miRNAs w/seed AAAGUGC),miR- 185-5p (and other miRNAs w/seed GGAGAGA),miR-200b-3p (and other miRNAs w/seed AAUACUG),miR-21-5p (and other miRNAs w/seed AGCUUAU),miR-221- 3p (and other miRNAs w/seed GCUACAU),miR-26a-5p (and other miRNAs w/seed UCAAGUA),miR-29b-3p (and other miRNAs w/seed AGCACCA) | 11 |
| 13 | Cell proliferation of hematoma cell lines | 0.00183 | Increased | 1.981 | let-7a-5p (and other miRNAs w/seed GAGGUAG),miR-100-5p (and other miRNAs w/seed ACCCGUA),miR-16-5p (and other miRNAs w/seed AGCAGCA),miR-29b-3p (and other miRNAs w/seed AGCACCA) | 4 |
| 14 | Cell proliferation of carcinoma cell lines | 0.000000136 | Increased | 2.004 | let-7a-5p (and other miRNAs w/seed GAGGUAG),miR-100-5p (and other miRNAs w/seed ACCCGUA),miR-143-3p (and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR- 155-5p (miRNAs w/seed UAAUGCU),miR- 17-5p (and other miRNAs w/seed AAAGUGC),miR-19b-3p (and other miRNAs w/seed GUGCAAA),miR-21-5p (and other miRNAs w/seed AGCUUAU),miR-24-3p (and other miRNAs w/seed GGCUCAG),miR-26a- 5p (and other miRNAs w/seed UCAAGUA) | 10 |
| 15 | Cell proliferation of tumor cell lines | 2.14E-09 | Increased | 2.346 | let-7a-5p (and other miRNAs w/seed GAGGUAG),miR-100-5p (and other miRNAs w/seed ACCCGUA),miR-125b-5p (and other miRNAs w/seed CCCUGAG),miR-130a-3p (and other miRNAs w/seed AGUGCAA),miR-143-3p (and other miRNAs w/seed GAGAUGA),miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-155-5p (miRNAs w/seed UAAUGCU),miR-16-5p (and other miRNAs w/seed AGCAGCA),miR-17-5p (and other miRNAs w/seed AAAGUGC),miR-185-5p (and other miRNAs w/seed GGAGAGA),miR-1908-5p (and other miRNAs w/seed GGCGGGG),miR-19b-3p (and other miRNAs w/seed GUGCAAA),miR-200b-3p (and other miRNAs w/seed | 21 |
| 16 | Invasion of breast cancer cell lines | 0.0000553 | Increased | 2.406 | miR-145-5p (and other miRNAs w/seed UCCAGUU),miR-155-5p (miRNAs w/seed UAAUGCU),miR-17-5p (and other miRNAs w/seed AAAGUGC),miR-200b-3p (and other miRNAs w/seed AAUACUG),miR-21-5p (and other miRNAs w/seed AGCUUAU),miR-29b-3p (and other miRNAs w/seed AGCACCA) | 6 |
Table 1. Differentially expressed miRNAs regulating various cellular and molecular functions in GC.
Conclusion
In conclusion, the novel GC-inducing miRNA signatures decrypted in our study can further be exploited for precision medicine in clinics. Our study may be helpful in future to identify potential prognostic biomarkers for GC in Saudi population.
Conflicts of interest
The authors declare no conflict of interest in the conduction of this study.
Acknowledgments
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH)–King Abdulaziz City for Science and Technology-the Kingdom of Saudi Arabia-award number (12-Bio-2725-03). The authors also, acknowledge with thanks Science and Technology Unit, King Abdulaziz University for technical support.
About the Authors
References
- Bibi F, Naseer MI, Alvi SA, Yasir M, et al. (2016). microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC Genom. 17:751. https://doi.org/10.1186/s12864-016-3090-7
- Feng JA (2016). Target research on tumor biology characteristics of mir-155-5p regulation on gastric cancer cell. Pak J Pharm Sci. 29:711-718.
- Feng Z, Zhang C, Wu R, Hu W. (2011). Tumor suppressor p53 meets microRNAs. J Mol Cell Biol.3:44-50. https://doi.org/10.1093/jmcb/mjq040
- Ghorai A, Ghosh U (2014). miRNA gene counts in chromosomes vary widely in a species and biogenesis of miRNA largely depends on transcription or post-transcriptional processing of coding genes. Front Genet. 5:100. https://doi.org/10.3389/fgene.2014.00100. eCollection 2014
- Heyns M, Kovalchuk O (2015). Non-coding RNAs including miRNAs, piRNAs, and tRNAs in human cancer. Oncotarget.6:23055-23057. https://doi.org/10.18632/oncotarget.5048
- Kramer A, Green J, Pollard J, Tugendreich S. (2014). Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics.30:523-530. https://doi.org/10.1093/bioinformatics/btt703
- Li H, Dai S, Zhen T, Shi H, et al. (2014). Clinical and biological significance of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer. 50:1207-1221. https://doi.org/10.1016/j.ejca.2013.12.010
- Liu L, Chen Q, Lai R, Wu X, et al. (2010). Elevated expression of mature miR-21 and miR-155 in cancerous gastric tissues from Chinese patients with gastric cancer. J Biomed Res. 24:187-197. https://doi.org/10.1016/S1674-8301(10)60028-0
- Lv QL, Du H, Liu YL, Huang YT, et al. (2017). Low expression of microRNA-320b correlates with tumorigenesis and unfavorable prognosis in glioma. Oncol Rep. 38:959-966. https://doi.org/10.3892/or.2017.5762
- Manikandan J, Aarthi JJ, Kumar SD, Pushparaj PN. Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2(8):330-4.
- Mei LL, Wang WJ, Qiu YT, Xie XF (2017). miR-125b-5p functions as a tumor suppressor gene partially by regulating HMGA2 in oesophageal squamous cell carcinoma. PLoS One. 12:e0185636. https://doi.org/10.1371/journal.pone.0185636
- Navarro F, Lieberman J. (2015). miR-34 and p53: New Insights into a Complex Functional Relationship. PLoSOne. 10:e0132767. https://doi.org/10.1371/journal.pone.0132767
- Palmero EI, de Campos SG, Campos M, de Souza NC, et al. (2011). Mechanisms and role of microRNA deregulation in cancer onset and progression. Genet Mol Biol. 34:363-370. https://doi.org/10.1590/S1415-47572011000300001
- Pekarsky Y, Croce CM. (2015). Role of miR-15/16 in CLL. Cell Death Differ. 22:6-11. https://doi.org/10.1038/cdd.2014.87
- Pushparaj PN, Aarthi JJ, Kumar SD, Manikandan J. (2008). RNAi and RNAa--the yin and yang of RNAome. Bioinformation. 2:235-237.
- Pushparaj PN, Aarthi JJ, Manikandan J, Kumar SD (2008). siRNA, miRNA, and shRNA: in vivo applications. J Dent Res. 87:992-1003. � https://doi.org/10.1177/154405910808701109
- Rong GQ, Zhang XM, Chen B, Yang XD, et al. (2017). MicroRNA gene polymorphisms and the risk of colorectal cancer. Oncol Lett. 13:3617-23. https://doi.org/10.3892/ol.2017.5885
- Shekari N, Baradaran B, Shanehbandi D, Kazemi T (2017). Circulating microRNAs: valuable biomarkers for diagnosis and prognosis of gastric cancer. Curr Med Chem. https://doi.org/10.2174/0929867324666171003123425
- Suarez-Arriaga MC, Torres J, Camorlinga-Ponce M, Gomez-Delgado A, et al. (2017). A proposed method for the relative quantification of levels of circulating microRNAs in the plasma of gastric cancer patients. Oncol Lett. 13: 3109-3117. https://doi.org/10.3892/ol.2017.5816
- Wang S, Han H, Hu Y, Yang W, et al. (2017). MicroRNA-130a-3p suppresses cell migration and invasion by inhibition of TBL1XR1-mediated EMT in human gastric carcinoma. Mol Carcinog. 2017. https://doi.org/10.1002/mc.22762
- Wu XL, Cheng B, Li PY, Huang HJ, et al. (2013). MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol.19:7758-65. https://doi.org/10.3748/wjg.v19.i43.7758
- Xie Y, Tobin LA, Camps J, Wangsa D, et al. (2013). MicroRNA-24 regulates XIAP to reduce the apoptosis threshold in cancer cells. Oncogene. 32:2442-51. https://doi.org/ 10.1038/onc.2012.258.
- Yang D, Zhan M, Chen T, Chen W, et al. (2017). MiR-125b-5p enhances chemotherapy sensitivity to cisplatin by down-regulating Bcl2 in gallbladder cancer.Sci Rep. 7:43109. https://doi.org/10.1038/srep43109
- You C, Liang H, Sun W, Li J, Liu Y, et al. (2017). Deregulation of the miR-16-KRAS axis promotes colorectal cancer. Sci Rep. 6:37459. https://doi.org/10.1038/srep37459
- Zhang Y, Guan DH, Bi RX, Xie J, (2017). Prognostic value of microRNAs in gastric cancer: a meta-analysis. Oncotarget. 8:55489-55510. https://doi.org/10.18632/oncotarget.18590
- Zhao LY, Tong DD, Xue M, Ma HL, et al. (2017). MeCP2, a target of miR-638, facilitates gastric cancer cell proliferation through activation of the MEK1/2-ERK1/2 signaling pathway by upregulating GIT1. Oncogenesis. 6:e368. https://doi.org/10.1038/oncsis.2017.60
- Zhao LY, Yao Y, Han J, Yang J, et al. (2014).� miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Dig Dis Sci. 59:1743-1753. https://doi.org/10.1007/s10620-014-3087-5
- Zheng L, Pu J, Qi T, Qi M, et al. (2013). miRNA-145 targets v-ets erythroblastosis virus E26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesis of gastric cancer cells. Mol Cancer Res. 11:182-93. https://doi.org/10.1158/1541-7786.MCR-12-0534
- Zhou J, Zhang M, Huang Y, Feng L, et al. (2015). MicroRNA-320b promotes colorectal cancer proliferation and invasion by competing with its homologous microRNA-320a. Cancer Lett. 356(2 Pt B):669-75. https://doi.org/10.1016/j.canlet.2014.10.014
- Zhou K, Liu M, Cao Y (2017). New Insight into microRNA Functions in Cancer: Oncogene-microRNA-Tumor Suppressor Gene Network. Front Mol Biosci. 4:46. https://doi.org/10.3389/fmolb.2017.00046
- Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, et al. (2010). MicroRNA-23a promotes the growth of gastric adenocarcinoma cell line MGC803 and downregulates interleukin-6 receptor. FEBS J. 277:3726-34. https://doi.org/10.1111/j.1742 4658.2010.07773.x
Keywords:
Download:
Full PDF- Share This