Cadmium accumulation and metallothionein gene expression in the liver of swamp eel (Monopterus albus) collected from the Mae Sot District, Tak Province, Thailand
Received: June 12, 2017
Accepted: August 11, 2017
Published: September 21, 2017
Genet.Mol.Res. 16(3): gmr16039748
DOI: 10.4238/gmr16039748
Abstract
Cadmium (Cd) is produced mainly as a by-product of zinc mining. In Thailand, the largest zinc mine is located in the Mae Sot district, Tak Province. Samples of Monopterus albus were collected from paddy fields in 4 sites, three downstream and one upstream from the zinc mine. The upstream site was considered to be uncontaminated while the three downstream sites were considered to be contaminated with Cd. Studies on the accumulation level of cadmium were conducted on the liver of the fish using the atomic absorption spectrophotometer technique. The metallothionein (MT) gene expression level in the liver, as a potential biomarker for long-term Cd exposure in their natural habitat, was also assessed. The level of hepatic MT gene expression was performed by quantitative real-time PCR. The result showed that Cd accumulation in the liver was much higher in swamp eels collected from the downstream sites when compared to those collected from the upstream site. The hepatic MT level in the upstream site was 0.75-fold, while the other three downstream sites were 0.36-, 4.44- and 0.94-fold. There is no parallel correlation between hepatic cadmium levels and hepatic MT gene expression. This study then suggests that MT gene expression biomarkers might be not suitable for swamp eels with prolonged exposure to Cd.
Introduction
Cadmium (Cd) is a heavy metal with no known biological function (Canli and Atli, 2003). It is one of the major stressors in aquatic systems in consequence of its ability to accumulate in living things over time (Ivanina et al., 2008). The major source of Cd discharge into the aquatic system is from non-ferrous metal mines. Cd contamination can be the result of mine drainage, waste water from ore processing, overflow from tailing reservoirs, and the run-off of rainwater from the area surrounding the mine. Thus, effluent flowing into local creeks and other water sources lead to extensive contamination downstream of the mining operation (IPCS, 1992). In freshwater fish, Cd could be taken from the environment in three possible ways, i.e., gills, skin, and intestines through ingestion (Jalaludeen et al., 2012). The essential metals such as copper, zinc, and iron, which are required for fish metabolism, must be obtained from the environment (water, food, or sediment). However, non-essential metals such as mercury, cadmium, and lead were found to be taken up via a similar route as essential ones thus leading to accumulation in fish tissue (Canli and Atli, 2003). Upon uptake, the metals are then delivered to the internal organs via the blood. In the blood plasma, various proteins participate in metal binding and, following transport, to internal organs for utilization, storage, and excretion (Chowdhury et al., 2003). After rapid clearance from the blood, Cd accumulates in several tissues in which the liver and kidneys tend to concentrate in higher levels the total content Cd of the fish body. The accumulation of cadmium in these tissues is probably due to their ability to synthesize the vast amount of metallothionein (Waalkes, 2000). Metallothionein (MT) is a low molecular weight cysteine-rich metal cytosolic binding peptide, which plays an important role in the homeostatic of essential metals, such as zinc and copper, and the detoxification of non-essential metals, such as cadmium and lead (Cheung et al., 2004). In the liver, Cd binds to already present MT by displacing zinc. Cd then induces the synthesis of new MT and then binds to it, thus protecting liver tissue from Cd toxicity (Nordberg and Nordberg, 2009). When the liver reaches saturation of Cd, Cd-MT complexes are then released into the blood stream and are delivered to the kidneys as the main storage site of Cd (Klaassen et al., 2009). MTs are strongly induced by heavy metals such as Cd; therefore, they are considered as a biomarker of metal exposure (Wangsongsak et al., 2007; Ivanina et al., 2008). The remarkable characteristic of all MTs in the protein level is the presence of Cys-Cys, Cys-X-Cys, and Cys- X-X-Cys structural motifs, where X is the amino acid other than Cys (Wang et al., 2014). The largest zinc mineral source in Thailand and also an area with high levels of Cd contamination is located in the Mae Sot District, Tak Province (Thamjedsada and Chaiwiwatworakul, 2012). Many studies have reported concerns regarding the contamination of Cd in this area. Simmons et al. (2005) found that soil samples obtained from rice field within Phratat Padaeng Sub District, Mae Sot, contain Cd in the range of 0.5 to 284 mg/kg. It is up to 114 times the Thai Investigation Level for Cd in soils. The rice grain in this area was also found to contain cadmium in the range of 0.05 to 7.7 mg/kg, exceeding the permissible level of Cd in rice, i.e., 0.2 mg/kg. Krissanakriangkrai et al. (2009) also reported that sediment and swamp eels in the Mae Tao River contain Cd that exceeds the maximum level allowed in sediment and fish. When fish are consumed by humans with high concentrations of Cd, around 3-7% of the ingested Cd is absorbed (Liu et al., 2015). Therefore, the level of cadmium in aquatic organisms such as fish must be given attention. The swamp eel, Monopterus albus, is one of the more common fish consumed by people in Mae Sot District. It is a predator and scavenger in freshwater ecosystems, thus can accumulate more contaminants (Krissanakriangkrai et al., 2009). The present study investigated Cd accumulation in the liver as well as MT gene expression levels in the liver of the swamp eel. This study also examined the relationship between Cd accumulation in liver and hepatic MT gene expression.
Materials and Methods
Study area and sampling collection
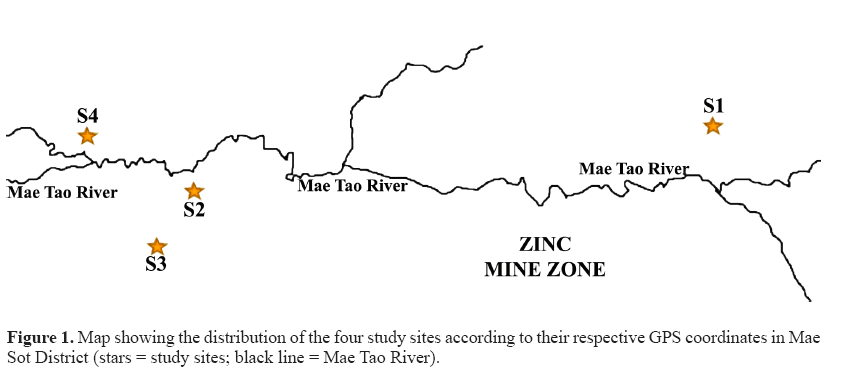
Thirty-two M. albus specimens were collected in July-August 2016 from four different paddy fields around the Mae Tao River, Mae Sot District, Tak Province, Thailand. The zinc mines are situated uphill of this river, along with the river path (Khaokaew and Landrot, 2015). The paddy fields have been receiving water irrigation from the Mae Tao River, which passes through the zinc mine area (Simmons et al., 2005). The Mae Tao River is regarded as the most worrying site for Cd contamination in Thailand (Netpae et al., 2015). Site S1 (16°0.40'36.55''N, 0.98°0.41'18.54''E) is located in Phratat Padaeng sub-district and was considered as an uncontaminated or less polluted site as it is located upstream and far from the zinc mine and the river. Meanwhile, sites S2, S3, and S4 were considered to be Cd contaminated and are located downstream from the zinc mine. Site S2 (16°0.39' 59.14''N, 0.98°0.36'56.19''E) and S3 (16°0.39'32.04''N, 0.98°0.36'38.31''E) are situated in Phratat Padaeng sub-district while site S4 (16°0.40'25.29''N, 098°0.36'01.37''E) is situated in Mae Tao sub-district. The study site is approximately 4-7 km downstream and is around 2-3 km upstream from the Zn mineralization zone (Figure 1).
The individual samples were collected from each location and brought to the laboratory for analysis and their size varied in a range between 70-110 g. The fish samples were then euthanized using clove oil and then carefully cut open using sharp scissors. The livers were taken for metal analysis using atomic absorption spectrophotometer (AAS) Perkin Elmer. Slices of the livers were also taken for the MT gene expression analysis. After removing the livers, they were submerged in RNAlater (Ambion, USA) immediately to stabilize the RNA and kept at -20°C until used. Protocols for the animal experiments were approved by the Naresuan University Animal Ethics Committee with registration number NU-AQ590201.
Cadmium level analysis of the liver
The liver tissues of 2 swamp eels from each site were pooled to make a subsample (replicate). Four replicates were taken from each site. The livers were dried in an oven at 150°C to constant weight. The samples were ground, and 0.5 g of each dried tissue was placed into polyethylene tubes. Five milliliters of freshly prepared nitric acid-perchloric acid (10:4) was added to the sample and allowed to digest overnight at room temperature. Final digestion was done by putting the digestion tubes in a water bath and set to 100°C and then boiled for about 2 h until all the tissues were dissolved. The samples were cooled to room temperature, filtered, transferred to 25-mL volumetric flasks and deionized water was added up to mark (Al-Weher, 2008). The digest solutions were transferred to plastic bottles before being analyzed using an AAS. According to Al-Weher (2008), determination of any heavy metals including cadmium in the sample can be calculated using the formula:
Real concentration of the metal in the sample (mg/kg) = ppmR x dilution factor where ppmR = reading solution by AAS (ppm) and dilution factor = volume of solution (mL) / weight of the dissolved sample (g).
Fish RNA isolation, synthesis of the first-strand cDNA, 3'-RACE amplification cDNA, cloning, and sequence analysis
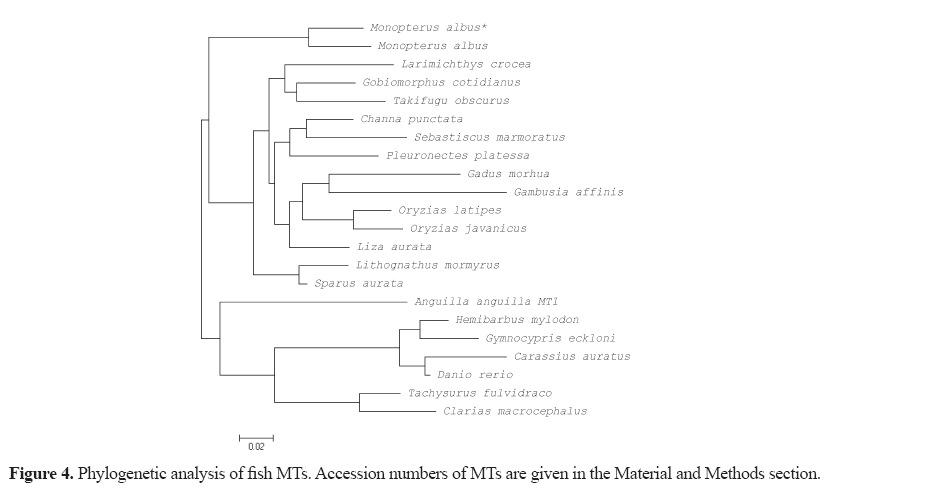
Total RNA from the liver tissue was isolated by using the TRIzol reagent (Ambion) according to the manufacturer’s protocol. Five micrograms of RNA was reverse transcribed with Tetro cDNA synthesis kit™ (Bioline, USA) using a dT-UPM primer (5'-AAGCAGTGGTATCAACGCAGAGTACTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT VN-3'). A gene specific primer was designed from a 32-sequence of the MT gene of fish in GenBank (NCBI). The 3'-rapid amplification of cDNA ends (3'- RACE) was performed to obtain the MT sequence. The Gene-specific primer (5'-ATGGAYCCYTGYGANTGCKCCAA-3') was used in conjunction with adaptor primer, UPM 1 (5'-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3'), to amplify 5'- and 3'-cDNA ends. The amplification was performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Lithuania) with a reaction mixture consisting of 4 μL 5X Phusion buffer, 0.2 μL Phusion DNA polymerase, 0.4 μL 10 mM dNTPs, 2 μL of each 10 μM UPM-1 and gene specific primer, 2 μL cDNA template and nuclease-free water to adjust to 20 μL. The polymerase chain reaction (PCR) condition was as follows: an initial denaturation for 2 min and 30 s at 98°C, followed by 35 cycles of 15 s at 98°C for denaturation, 15 s at 60°C for annealing, 15 s at 72°C for extension and 5 m at 72°C for a final extension. The predicted size of PCR products was purified using RBC Real Genomics Hi Yield Gel/PCR DNA fragment extraction kit (RBC Bioscience, Taiwan) and, then, ligated into the pJET1.2/ blunt cloning vector (Thermo Scientific, USA) following transformation into Escherichia coli strain JM109 using the heat shock method. Transformed colonies were screened byperforming PCR using primers under the same PCR conditions. The plasmid extraction was performed using GeneJET Plasmid Miniprep Kit (Fermentas, USA) and was sequenced by Macrogen (Korea) . The sequences were then aligned using the Clustal Omega program (EMBL-EBI, UK) to obtain the MT gene sequence of M. albus. Protein prediction was performed using the ExPASy program (Swiss Institute of Bioinformatics) and compared to other reported sequences using BLAST (NCBI) to identify similarity. Multiple sequence alignments of deduced amino acid sequences and other reported fish MT sequences were generated using MEGA 5.05. M. albus and reported fish MT amino acid sequences were used to construct a neighbor-joining (N -J) phylogenetic tree using MEGA 5.05. The accession number in the GenBank (NCBI) of the other reported fish MTs are as follows: M. albus (XP_020460040.1), Larimichthys crocea (AIT55913.1), Gobiomorphus cotidianus (AEO52916.1), Channa punctata (ACL31528.1), Sebastiscus marmoratus (JQ389121.1), Pleuronectes platessa (CAA40067.1), Gadus morhua (CAA65924.1), Oryzias latipes (NP_001098255.1), Oryzias javanicus (AAW83513.2), Lithognathus mormyrus (AF321007.1), Sparus aurata (U58774.1), Liza aurata (AAB51591.1), Gambusia affinis (AB455145.1), Takifugu obscurus (ABU94445.1), Anguilla anguilla MT 1 (DQ493910), Hemibarbus mylodon (ABS87376.1), Gymnocypris eckloni (KC352712.1), Carassius auratus (AAB32777.1), Danio rerio (AY514790.1), Tachysurus fulvidraco (ABW88898.1),and Clarias macrocephalus (JX312865.1).
Real-time PCR
Total RNA isolation cDNA synthesis
Two micrograms of isolated RNA from liver was reverse transcribed with Tetro cDNA synthesis kitTM (Bioline). The cDNA mixture was stored at -20°C until used in the real-time PCR.
Quantitation of MT gene expression by real-time PCR
Amplification and quantitation of cDNA were performed in the Swift Spectrum™ 48 Real-Time Thermal Cycler (ESCO Micro Pte. Ltd., Thailand). The primers of the MT gene were designed from the conserved region of previously cloned MT sequence. Meanwhile, the primers of the 18S rRNA gene were selected according to Hu et al. (2014). The 18S rRNA gene of the swamp eel served as an internal control for normalization and samplesfrom site S1 (reference site) served as a calibrator. The relative gene expression data were calculated using comparative Ct method (2-â�?�?â�?�?Ct method) as described previously by Livak and Schmittgen (2001). Amplification was carried out in a final volume of 20 µL containing 10 µL 2X SensiFAST SYBR (Bioline), 0.3 µL of each 10 µM forward and reverse primers, and 8.4 µL nuclease-free water. The primers of MT and 18S rRNA sequence for real-time PCR are listed in Table 1. The real-time PCR condition was as follows: 95°C for 3 min, followed by 40 cycles of 15 s at 95°C, 15 s at 58°C, 15 s at 72°C. Each sample was run in triplicate and mean values were reported.
| Primer | Sequences | PCR product (bp) |
| 18S rRNA | F 5'-GTGGAGCGATTTGTCTGGTTA-3' | 162 |
| R 5'-CGGACATCTAAGGGCATCAC-3' | ||
| MT | F 5'-CTGCTCATGTTGTCCTTCCG-3' | 70 |
| R 5'-GTCGCACTTCTTCCCTTTGC-3' |
Table 1: Primer sequences of 18S rRNA and metallothionein (MT) gene for real-time PCR.
Statistical analysis
All data are reported as means ± standard error (means ± SE) value. One-way ANOVA was used for determining the significant difference in Cd and MT gene expression levels followed by Duncan multiple range tests for statistical comparison in each experiment. The Spearman correlation test was applied to study the correlation between hepatic Cd levels and hepatic MT levels. Significance is accepted for P < 0.05. Statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Cd accumulation in the liver
The result of Cd accumulation in livers from 4 different sampling sites is summarized in Table 2. The Cd level differed significantly between site S4 and the other three sites (S1, S2, and S3). Significant different of Cd level was also found between site S2 and site S1. The highest liver Cd levels were found in site S4 while the lowest were found in site S1. In general, the fish liver collected from the three downstream sites accumulated more Cd when compared to the fish liver collected from the upstream site. The trend of liver Cd accumulation according to the site was S4 > S2 > S3 > S1.
| Location (sub-district) | Code | Liver |
| Means ± SE | ||
| Phratat Padaeng | S1 | 1.10 ± 0.10c |
| Phratat Padaeng | S2 | 10.26 ± 3.04b |
| Phratat Padaeng | S3 | 6.70 ±1.26bc |
| Mae Tao | S4 | 22.88 ± 2.60a |
Means of Cd level having a different letter are significantly different, P < 0.05.
Table 2: Accumulation of Cd in Monopterus albus liver (mg/kg dry weight) from the four study sites.
M. albus MT cloning characterization
The MT mRNA sequence of 333 bp, which consists of a complete coding sequence and 3'-untranslated region (3'-UTR), was identified from swamp eel (M. albus). The MT cDNA sequence obtained from M. albus consists of a coding region in length of 180 bp encoding 60 amino acids with 20 cysteine residues and a 3'-UTR of 153 bp with a poly(A) tail. The βâ�?�?domain of M. albus MT protein comprises 9 cysteine residues and 11 cysteine residues in the α α domain. The sequence contains Cys-X-Cys, Cys-XX-Cys, and Cys-Cys pattern (Figure 2), which is the standard distribution of cysteine residues in this protein.
Comparison of M. albus and other fish MTs and phylogenetic analysis
The alignment of the amino acid sequence of M. albus MT shows similarity (identity) with other fish MTs ranging from 77 to 93% (Figure 3). The MT gene of M. albus comprises 33% cysteine residues where their positions were found to be highly conserved among fish MTs. The amino acid comparison between M. albus and the other reported fish MTs shows the variation of amino acid at positions 5 (E-D or A), 7 (S-A), 9 (T-S), 11 (T-N or S or K or A), 13 (N-T or K or S), 16 (A-G or T or D), 17 (N-S or T), 19 (S-N or T or K or A), 21 (K-T or A or S), 22 (D-N or S or G), 24 (S-K or Q), 26 (T-K), 27 (T-S or K), 29 (K-N), 34 (S-P or E or A or F), 38 (S-A), 39 (G-D or T), 41 (S-T or P), 45 (A- S), 50 (K-N), 51 (G-D), 52 (K-E or D or N), 53 (K-T or S), 55 (D-G), 56 (T-K or P or S or A), and 57 (S-T or N or G or K).
Figure 3: Multiple sequence alignments of the Monopterus albus* MT protein with the other reported fish MT proteins. The percentages of identities are displayed on the right side of the species name. The asterisk indicates the positions of amino acids that have fully conserved residue. Accession numbers of MTs are given in the Material and Methods section.
The protein sequences of M. albus and other fish MTs were subjected to phylogenetic analysis, which then shows the evolutionary conservation of various fish MTs and how distantly related they are (Figure 4).
Hepatic MT gene expression
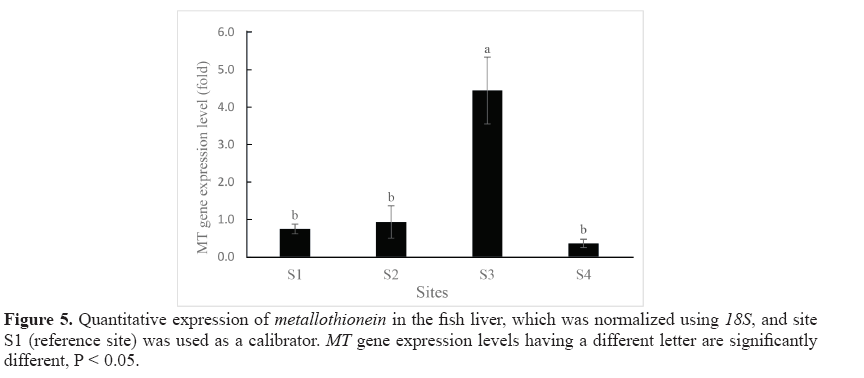
Expression levels of MT in the liver of swamp eels collected from the 4 study sites are shown in Figure 5. By using site S1 as the calibrator, it was shown that the hepatic MT gene was expressed in the samples from all study sites. Statistically significant differences in MT gene expression level were found between site S3 and the other three sites (S1, S2, and S4). The highest level MT mRNA expression was found in site S3 while the lowest level was found in site S4.
Correlation liver Cd-MT gene expression
Spearman’s correlation coefficient showed moderate negative correlation and was not significant (R = -0.400, P > 0.05) between hepatic MT mRNA level and the hepatic Cd content. The hepatic MT gene expression levels did not follow the trend of Cd level in the liver. For example, hepatic Cd content from site S3 and site S2 showed no significant difference (P > 0.05, 6.70 ± 1.26 and 10.26 ± 3.04 mg/kg dw, respectively), while MT gene expression levels on these two sites were significantly different (P < 0.05, 4.44 ± 0.89 and 0.94 ± 0.43-fold of MT mRNA, respectively). Contrarily, no significant differences in MT gene expression was found between S1 and S4 (P > 0.05, 0.75 ± 0.13 and 0.36 ± 0.11-fold of MT mRNA, respectively) whereas the hepatic Cd levels from these two sites were the lowest and the highest and showed significant difference (P < 0.05, 1.10 ± 0.10 and 22.66 ± 2.60 mg/kg dw, respectively). Similar result was also found between site S1 and site S2 where the MT gene expression levels were not significantly different (P > 0.05, 0.75 ± 0.13 and 0.94 ± 0.43-fold, respectively) while the hepatic Cd levels from these two sites were significantly different (P < 0.05, 1.10 ± 0.10 and 10.26 ± 3.04 mg/kg dw, respectively).
Discussion
The current study was carried out around Mae Tao Creek in four different locations. M. albus is one of the most common fish that can be found in the paddy fields; therefore, it has beensuggested as an object of study for monitoring heavy metal pollution in the paddy fields (Yin et al., 2012). This study revealed that the average Cd accumulation in the liver of swamp eels differed depending on the location. It showed that the swamp eel obtained from downstream sites (site S2, S3, and S4) accumulated much higher cadmium levels in the liver compared to those from the upstream site (site S1), indicating that the downstream site environment might be contaminated with more Cd compared to the upstream site. This view is supported by the previous study by Weeraprapan et al. (2015) who found that the Cd levels in the sediment of Mae Tao Creek, which is located downstream of the zinc mine, were much higher than in the sediment collected from upstream of the zinc mine areas. As the Mae Tao River accepts water from the zinc mine, it is apparent that the source of Cd is the zinc mine (Weeraprapan et al., 2015). This is also in line with Thamjedsada and Chaiwiwatworakul (2012) who reported that Cd in the sediment of Mae Tao Creek tends to be higher in downstream sites from the zinc mine both in the dry and the wet seasons. Since site S1 is located far above the zinc mine, it receives irrigation water, which has not passed the zinc mine area, thus minimizing Cd contamination. Sites S2, S3, and S4, on the other hand, receive water from the river, which passes through the zinc mine compound that is highly contaminated with Cd. Some mining activities such as drilling, material transfer, the removal of mine tailings, and drainage may influence the contamination of the environment with Cd (Netpae et al., 2015). Cd is classified as a heavy metal, which is non-biodegradable and non-thermodegradable, and hence it readily accumulates to toxic levels (Taghipour et al., 2012). This heavy metal can then cause heavy metal pollution in bodies of water through deposition and is incorporated in water, sediment, and aquatic life (Abdel-Baki et al., 2011). Apart of being located downstream from the zinc mine, for the downstream sites, the distance of the sites from the Mae Tao River seems to affect the Cd accumulation levels in the liver of the swamp eels. The liver accumulated Cd at the highest levels in sites S4 and S2, as these sites were closest to the Mae Tao River (Figure 1) 1) while site S3 has a lower level of hepatic Cd as it is located farther from Mae Tao River than the other two sites.
In the current study, the MT gene sequence was cloned and identified from the liver of swamp eel. Several features in M. albus MT protein sequences showed the typical characteristics of fish MT; for instance, the length of MTs, in which most of the teleost MTs contains 60 amino acids, except for metallothionein A from rainbow trout and Atlantic salmon, which contain 61 amino acids, and also for white sturgeon and lake sturgeon, which contain 63 amino acids (Doering et al., 2015). In mammalian MT, the last Cys-X-Cys-Cys motif in α domain becomes Cys-XXX-Cys-Cys in fish MT, which marks the difference between mammalian and fish MT protein sequences (D’Auria et al., 2001). This protein also shows another typical MT characteristic such as the absence of aromatic amino acid and the presence of 5 glycine residues (Ceratto et al., 2002). Multiple sequence alignments and phylogenetic tree analysis of our MT M. albus protein and other known fish MT protein showed that the protein shared a high similarity with other fishes.
The MT gene expression of swamp eel liver was then determined quantitatively using real-time PCR. The most intense MT synthesis is usually observed in the liver, which is probably related to its function as a major detoxification organ (Kovarova et al., 2009). Yang et al. (2014) also noted that the MT gene was detected in all eleven tissues (gills, heart, skin, spleen, head kidney, blood, kidney, stomach, intestine, brain, and liver) of healthy black porgy, indicating that MT mRNA was ubiquitous in various tissues; however, liver has the highest MT gene expression. The liver was thus chosen as the subject organ in the present study. In thepresent study, the MT expression level was used in an attempt to understand its suitability as a biomarker of metal exposure (especially Cd) as it is frequently used in pollution monitoring programs (Quirós et al., 2007). Hepatic MT gene expression was observed in all study sites including uncontaminated sites and/or in the lowest hepatic Cd level samples, implying that Cd detoxification is not the only role of MT and/or that the constitutive expression of MT is a basal level of protection from Cd toxicity (Timmermans et al., 2005). It has previously been suggested by Bourdineaud et al. (2006) who found that under low contamination stress, the purpose of the MT gene induction was to compensate MT degradation rather than to increase metal sequestration capacities as expected. This is suitable for the role of MT gene induction regarding zinc homeostasis rather than to metal detoxification. Metallothionein is expected to be induced under conditions of elevated metal concentrations, allowing more binding sites to sequester the metal ions and preventing potential adverse effects (Langston et al., 2002). Some studies in a variety of fish such as killifish (Van Cleef-Toedt et al., 2001), silver barb (Wangsongsak et al., 2007) and grass carp (Tan et al., 2016) showed the elevation of MT expression level after being exposed to Cd. In the current study, the highest MT geneexpression of the liver was found in site S3 of 5.92-fold followed by site S2 of 1.25-fold compared to site S1 (reference site). Earlier field studies have shown that hepatic MT gene expression of Oreochromis niloticus was found to be higher in heavy metal polluted fish farms (Abumourad et al., 2013). Interestingly, the present study showed no parallel correlation between MT gene expression levels and hepatic cadmium content. The current study revealed that the lowest expression of 0.48-fold (compared to S1) was found in S4, which turned out to be the highest hepatic Cd level site. This is in agreement with the findings of Shariati et al. (2011) who found that the Cd levels in the liver of Persian sturgeon increased as the Cd concentration and time exposure increased; however, MT protein level increased only in short period (4 days) after being exposed to Cd, and there was no significant change of MT
protein level between 4- and 14-day treatment groups. The accumulations of Cd in the liver might exceed its regulation capacity, leading to disruption in the induction of MT synthesis and eventually loss of the correlation between MT and Cd levels as found at lower exposed concentrations (Shariati et al., 2011). When the metal level has surpassed the capability of an animal to produce MT, the metal may bind to high molecular mass proteins, but with a weaker affinity than the Cd toxicity may follow (De Boeck et al., 2003). For this reason, MT gene expression biomarker was probably more sensitive and suitable for acute (short-term) exposure but not for chronic (long-term) exposure to heavy metals (Quirós et al., 2007; Rhee et al., 2009). However, in other species such as springtail, the animal might develop Cd tolerance after chronic exposure by increasing the basal level of MT gene expression. It was proved by the higher expression of MT basal levels of this animals’ population collected from a metal-contaminated environment, compared to those obtained from a clean environment, even after the animals have been cultured in a laboratory with clean food for several generations (Timmermans et al., 2005). Earlier laboratory studies by Kovarova et al. (2009), in which the hepatic MT protein in carp reached its maximum level (>130 ng/g) in lower exposure to Cd (2.5, 5, 7.5 mg/L CdCl) yet hit the low MT levels (<50 ng/g) in conditions with the highest exposure to Cd (12.5 mg/L CdCl) and control groups (Cd-free treatment) in all time periods of exposure (24, 48, 72, and 96 h). Wangsongsak et al. (2007) also demonstrated that during 56 days of exposure of silver barb to three series amounts of Cd, i.e., 0.012, 0.06, and 0.12 mg/L, consecutively, the hepatic and renal MT mRNA expression increased significantly upon the exposure and reached its highest expression at 28 days. However, MT expressions both in liver and kidney gradually dropped after 35 days. Hepatic MT and renal MT expressions remain high and low, respectively, after fish removal to Cd-free water for 4 weeks. The induction and synthesis of MT and its binding capacity are restricted (Kovarova et al., 2009). This matter has also been observed in human monocytes exposed to CdCl that showed that 10 µM Cd2+, which induced significant toxicity, induced significantly less MT mRNA than 1 µM Cd2+. It was then thought that MT levels perhaps have to be in a relatively narrow range to mediate the induction of activation; either increasing or decreasing those levels would cause negative effects. Another possible hypothesis that has been proposed is that heavy metal exposure might affect the other expression of genes in addition to MT’s induction. The non-MT factors induced by metal might act on their own or in concert with MT to inhibit theactivation potential (Koropatnick and Zalups, 1997). MT was found to be strongly induced by Cd in the laboratory (Wangsongsak et al., 2007) and field condition (Fernandes et al., 2008); however, not every species of fish is suitable for bio-monitoring (Kovarova et al., 2009). It was reported that MT was suitable as a bioindicator for monitoring heavy metal pollution in brown trout, but not in European eel (Linde et al., 2001). In animals, it is thought that the increase of susceptibility to Cd toxicity is usually associated with the inability of the animal to synthesize MT (W�?�?ostowski et al., 2008). Wu et al. (2006) observed that three species of fish, i.e., Acrossocheilus paradoxus, Oreochromis mossamibicus, and Chanos chanos, appear to have a different tolerance to Cd. It is possibly due to different Cd accumulation rates and the ability to synthesize the supplementary MT in each species (Wu et al., 2006). Furthermore, several stressors - other than metal - have been reported to predispose the MT mRNA synthesis. Yang et al. (2014) reported a significant increase in MT gene expression in black porgy after 48 h of bacterial infection, suggesting that the MT gene in this species is involved in the immune-associated response. Abdel-Tawwab and Wafeek (2014) observed that the temperature significantly increases the MT levels in respect to the increase in Cd
accumulation. Rhee et al. (2009) indicated that some significant factors, such as age and gender, were found to affect the MT gene expression level of Kryptolebias marmoratus (Mangrove killifish). It is worth noting that, with swamp eels, it was not always possible to distinguish the gender or estimate the age precisely. Besides, the variation of swamp eel size in this study may be related to the variation of accumulated Cd thus affecting the MT gene expression level. Even though, MT mRNA expression can be stimulated by various stressors, hepatic MT in eel (Anguilla anguilla) was found to be strongly correlated with hepatic heavy metal, even after taking other variables (sizes and seasonal variations) into consideration, indicating that the MT is a good biomarker for heavy metal contamination (Bird et al., 2008).
Conclusion
From this study, we concluded that high hepatic Cd levels in downstream sites were attributed to the Cd accumulation in swamp eels living in paddy fields irrigated with Cd-contaminated water from the upstream zinc mine. This study suggests that Cd may induce MT gene expression within certain range levels only, which could be due to the prolonged duration of exposure and high concentration exposure to Cd. Cd accumulation might have exceeded the limit of its regulatory capacity resulting in the restriction of MT mRNA synthesis. Furthermore, some intrinsic and extrinsic factors need to be taken into consideration. Therefore, before the application of the MT gene expression biomarker in the natural environment, it is important to develop laboratory studies to provide data regarding the basal and maximum induced levels of swamp eel MT mRNA and also environmental variables that may regulate MT gene expression in this species.
Acknowledgments
The authors are very grateful to the Naresuan University for providing financial support (grant #R250C082). We also thank Mr. Kevin Mark Roebl in the Writing Clinic of Naresuan University Language Center (NULC) for improvement and English correction.
About the Authors
Corresponding Author
N. Yimtragool
Department of Biology, Faculty of Science, Naresuan University, Phitsanulok, Thailand
- Email:
- nonglakp@nu.ac.th
References
- Abdel-Baki AS, Dkhil MA and Al-Quraishy S (2011). Bioaccumulation of some heavy metals in tilapia fish relevant to their concentration in water and sediment of Wadi Hanifah, Saudi Arabia. Afr. J. Biotechnol. 10: 2541-2547.
- Abdel-Tawwab M and Wafeek M (2014). Influence of water temperature and waterborne cadmium toxicity on growth performance and metallothionein-cadmium distribution in different organs of Nile tilapia, Oreochromis niloticus (L.). J. Therm. Biol. 45: 157-162. https://doi.org/10.1016/j.jtherbio.2014.09.002
- Abumourad IMK, Authman MMN and Abbas WT (2013). Heavy metal pollution and metallothionein expression: a survey on Egyptian tilapia farms. J. Appl. Sci. Res 9: 612-619.
- Al-Weher SM (2008). Levels of heavy metal Cd, Cu and Zn in three fish species collected from the Northern Jordan Valley, Jordan. JJBS 1: 41-46.
- Bird DJ, Rotchell JM, Hesp SA, Newton LC, et al. (2008). To what extent are hepatic concentrations of heavy metals in Anguilla anguilla at a site in a contaminated estuary related to body size and age and reflected in the metallothioneinconcentrations? Environ. Pollut. 151: 641-651. https://doi.org/10.1016/j.envpol.2007.03.018
- Bourdineaud JP, Baudrimont M, Gonzalez P and Moreau JL (2006). Challenging the model for induction of metallothionein gene expression. Biochimie 88: 1787-1792. https://doi.org/10.1016/j.biochi.2006.07.021
- Canli M and Atli G (2003). The relationships between heavy metal (Cd, Cr, Cu, Fe, Pb, Zn) levels and the size of six Mediterranean fish species. Environ. Pollut. 121: 129-136. https://doi.org/10.1016/S0269-7491(02)00194-X
- Ceratto N, Dondero F, van de Loo JW, Burlando B, et al. (2002). Cloning and sequencing of a novel metallothionein gene in Mytilus galloprovincialis Lam. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 131: 217-222. https://doi. org/10.1016/S1532-0456(02)00008-X
- Cheung AP, Lam TH and Chan KM (2004). Regulation of Tilapia metallothionein gene expression by heavy metal ions. Mar. Environ. Res. 58: 389-394.https://doi.org/10.1016/j.marenvres.2004.03.084
- Chowdhury MJ, Grosell M, McDonald DG and Wood CM (2003). Plasma clearance of cadmium and zinc in non-acclimated and metal-acclimated trout. Aquat. Toxicol. 64: 259-275. https://doi.org/10.1016/S0166-445X(03)00074-2
- D’Auria S, Carginale V, Scudiero R, Crescenzi O, et al. (2001). Structural characterization and thermal stability of Notothenia coriiceps metallothionein. Biochem. J. 354: 291-299.https://doi.org/10.1042/bj3540291
- De Boeck G, Ngo TT, Van Campenhout K and Blust R (2003). Differential metallothionein induction patterns in three freshwater fish during sublethal copper exposure. Aquat. Toxicol. 65: 413-424. https://doi.org/10.1016/S0166-445X(03)00178-4
- Doering JA, Beitel SC, Eisner BK, Heide T, et al. (2015). Identification and response to metals of metallothionein in two ancient fishes: white sturgeon (Acipenser transmontanus) and lake sturgeon (Acipenser fulvescens). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 171: 41-48.https://doi.org/10.1016/j.cbpc.2015.03.002
- Fernandes D, Bebianno MJ and Porte C (2008). Hepatic levels of metal and metallothioneins in two commercial fish species of the Northern Iberian shelf. Sci. Total Environ. 391: 159-167. https://doi.org/10.1016/j.scitotenv.2007.10.057
- Hu Q, Guo W, Gao Y, Tang R, et al. (2014). Reference gene selection for real-time RT-PCR normalization in rice field eel (Monopterus albus) during gonad development. Fish Physiol. Biochem. 40: 1721-1730. https://doi.org/10.1007/ s10695-014-9962-3
- IPCS (International Programme on Chemical Safety) (1992). Cadmium - Environmental Aspects. World Health Organization, Geneva.
- Ivanina AV, Cherkasov AS and Sokolova IM (2008). Effects of cadmium on cellular protein and glutathione synthesis and expression of stress proteins in eastern oysters, Crassostrea virginica Gmelin. J. Exp. Biol. 211: 577-586. https://doi. org/10.1242/jeb.011262
- Jalaludeen MD, Arunachalam M, Raja M, Nandagopal S, et al. (2012). Histopathological of the gill, liver and kidney tissue of the freshwater fish Tilapia Mossambica exposed to cadmium sulphate. IJABBR 2: 572-578.
- Khaokaew S and Landrot G (2015). A field-scale study of cadmium phytoremediation in a contaminated agricultural soil at Mae Sot District, Tak Province, Thailand: (1) Determination of Cd-hyperaccumulating plants. Chemosphere 138: 883-887. https://doi.org/10.1016/j.chemosphere.2014.09.108
- Klaassen CD, Liu J and Diwan BA (2009). Metallothionein protection of cadmium toxicity. Toxicol. Appl. Pharmacol. 215-220. https://doi.org/10.1016/j.taap.2009.03.026
- Koropatnick J and Zalups RK (1997). Effect of non-toxic mercury, zinc or cadmium pretreatment on the capacity of human monocytes to undergo lipopolysaccharide-induced activation. Br. J. Pharmacol. 120: 797-806. https://doi. org/10.1038/sj.bjp.0700975
- Kovarova J, Kizek R, Adam V, Harustiakova D, et al. (2009). Effect of cadmium chloride on metallothionein levels in carp. Sensors (Basel) 9: 4789-4803.https://doi.org/10.3390/s90604789
- Krissanakriangkrai O, Supanpaiboon W, Juwa S, Chaiwong S, et al. (2009). Bioavailable cadmium in water and fish, in highly contaminated area on the Thai-Myanmay Border. Thammasat Int. J. Sci. Tech. 14: 60-68.
- Langston WJ, Chesman BS, Burt GR, Pope ND, et al. (2002). Metallothionein in liver of eels Anguilla anguilla from the Thames Estuary: an indicator of environmental quality? Mar. Environ. Res. 53: 263-293. https://doi.org/10.1016/ S0141-1136(01)00113-1
- Linde AR, Sánchez-Galán S, Vallés-Mota P and García-Vázquez E (2001). Metallothionein as bioindicator of freshwater metal pollution: European eel and brown trout. Ecotoxicol. Environ. Saf. 49: 60-63. https://doi.org/10.1006/ eesa.2001.2042
- Liu K, Chi S, Liu H, Dong X, et al. (2015). Toxic effects of two sources of dietborne cadmium on the juvenile cobia, Rachycentron canadum L. and tissue-specific accumulation of related minerals. Aquat. Toxicol. 165: 120-128.https://doi.org/10.1016/j.aquatox.2015.05.013
- Livak KJ and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(- C(T)) Method. Methods 25: 402-408. https://doi.org/10.1006/meth.2001.1262
- Netpae T, Suckley S and Phalaraksh C (2015). Cadmium tolerance fungi isolated from polluted sites in the Mae Tao creek, Thailand. Adv. Stud. Biol. 7: 29-37. https://doi.org/10.12988/asb.2015.41050
- Nordberg M and Nordberg GF (2009). Metallothionein: historical development and overview. In: Metallothionein and related chelators series metal ion in life science 5 (Sigel A, Sigel H and Sigel RKO, eds.). John Wiley & Sons, Ltd., Chichester.
- Quirós L, Piña B, Solé M, Blasco J, et al. (2007). Environmental monitoring by gene expression biomarkers in Barbus graellsii: laboratory and field studies. Chemosphere 67: 1144-1154. https://doi.org/10.1016/j.chemosphere.2006.11.032
- Rhee JS, Raisuddin S, Hwang DS, Lee KW, et al. (2009). Differential expression of metallothionein (MT) gene by trace metals and endocrine-disrupting chemicals in the hermaphroditic mangrove killifish, Kryptolebias marmoratus. Ecotoxicol. Environ. Saf. 72: 206-212.https://doi.org/10.1016/j.ecoenv.2008.06.001
- Shariati F, Esaili Sari A, Mashinchian A and Pourkazemi M (2011). Metallothionein as potential biomarker of cadmium exposure in Persian sturgeon (Acipenser persicus). Biol. Trace Elem. Res. 143: 281-291. https://doi.org/10.1007/ s12011-010-8877-9
- Simmons RW, Pongsakul P, Saiyasitpanich D and Klinphoklap S (2005). Elevated levels of cadmium and zinc in paddy soils and elevated levels of cadmium in rice grain downstream of a zinc mineralized area in Thailand: implications for public health. Environ. Geochem. Health 27: 501-511. https://doi.org/10.1007/s10653-005-7857-z
- Taghipour H, Mosaferi M, Pourakbar M and Armanfar F (2012). Heavy metals concentrations in groundwater used for irrigation. Health Promot. Perspect. 2: 205-210.
- Tan S, Li H, Jin Y and Yu H (2016). Cadmium exposure effects on the expression of metallothionein 2 gene in grass carp (Ctenopharyngodon idellus). Genes Genomics 38: 127-135. https://doi.org/10.1007/s13258-015-0347-5
- Thamjedsada T and Chaiwiwatworakul P (2012). Evaluation of cadmium contamination in the Mae Tao Creek sediment.ICSET 27: 501-511.
- Timmermans MJ, Ellers J, Roelofs D and van Straalen NM (2005). Metallothionein mRNA expression and cadmium tolerance in metal-stressed and reference populations of the springtail Orchesella cincta. Ecotoxicology 14: 727-739. https://doi.org/10.1007/s10646-005-0020-x
- Van Cleef-Toedt KA, Kaplan LAE and Crivello JF (2001). Killifish metallothionein messenger RNA expression following temperature perturbation and cadmium exposure. Cell Stress Chaperones 6: 351-359. https://doi.org/10.1379/1466-1268(2001)006<0351:KMMREF>2.0.CO;2
- Waalkes MP (2000). Cadmium carcinogenesis in review. J. Inorg. Biochem. 79: 241-244. https://doi.org/10.1016/S0162-0134(00)00009-X
- Wang WC, Mao H, Ma DD and Yang WX (2014). Characteristics, functions, and applications of metallothionein in aquatic vertebrates. Front. Mater. Sci. 1: 34.
- Wangsongsak A, Utarnpongsa S, Kruatrachue M, Ponglikitmongkol M, et al. (2007). Alterations of organ histopathology and metallothionein mRNA expression in silver barb, Puntius gonionotus during subchronic cadmium exposure. J. Environ. Sci. (China) 19: 1341-1348.https://doi.org/10.1016/S1001-0742(07)60219-8
- Weeraprapan P, Phalaraksh C, Chantara S and Kawashima M (2015). Water quality monitoring and cadmium contamination in the sediments of Mae Tao Stream, Mae Sot District, Tak Province, Thailand. IJESD 6: 142-146. https://doi. org/10.7763/IJESD.2015.V6.577
- W�?�?ostowski T, Krasowska A and Bonda E (2008). Joint effects of dietary cadmium and polychlorinated biphenyls on metallothionein induction, lipid peroxidation and histopathology in the kidneys and liver of bank voles. Ecotoxicol. Environ. Saf. 69: 403-410.https://doi.org/10.1016/j.ecoenv.2007.03.014
- Wu SM, Jong KJ and Lee YJ (2006). Relationships among metallothionein, cadmium accumulation, and cadmium tolerance in three species of fish. Bull. Environ. Contam. Toxicol. 76: 595-600. https://doi.org/10.1007/s00128-006-0961-7
- Yang M, Pan C, Bo J and Wang K (2014). Regulation of metallothionein gene expression in response to benzo[a]pyrene exposure and bacterial challenge in marine cultured black porgy (Acanthopagrus schlegelii). Chin. J. Geochem. 33: 404-410. https://doi.org/10.1007/s11631-014-0705-z
- Yin SA, Ismail A and Zulkifli SZ (2012). Heavy metals uptake by Asian swamp eel, Monopterus albus from paddy fields of Kelantan, Peninsular Malaysia: Preliminary Study. Trop. Life Sci. Res. 23: 27-38.
Keywords:
Download:
Full PDF- Share This