Assessment of allelic diversity among drought-resistant cotton genotypes using microsatellite markers
Published: May 25, 2017
Genet.Mol.Res. 16(2): gmr16029664
DOI: 10.4238/gmr16029664
Abstract
Drought, in conjunction with high temperature, is an important environmental constraint to cotton production. Development of cotton varieties with increased tolerance against adverse environmental conditions has been proposed as effective strategy for ensuring reliable yields. In the present study, 30 simple sequence repeat (SSR) primers were used to estimate genetic divergence among 22 cotton genotypes for drought stress tolerance. Genetic diversity is a prerequisite for developing drought resistant cotton genotypes. Eleven SSR primers out of 30 were able to discriminate among the cotton genotypes, implying that 37% of the primers were informative. In total, 41 alleles were detected, with an average of 3.72 alleles per primer. The number of alleles per locus ranged from one (JESPR-284) to six (JESSPR-302), and the allelic diversity in the experimental material was 0.40. Genetic similarity coefficients ranged from 0.87-1.00. The result of principal component analysis confirmed the clustering of 21 cotton genotypes in two groups leaving one genotype (CIM-109) ungrouped. Overall, genetic diversity among the 22 cotton genotypes was low. More polymorphic SSR markers are needed to explore the workable genetic variation among the screened cotton genotypes in future studies.
https://sporbahisleri.livejournal.com https://wakelet.com/@SporBahisleri67459 https://theomnibuzz.com/author/sporbahisleri/ https://lessons.drawspace.com/profile/322207/sporbahisleri/workflow https://writeupcafe.com/profile/sporbahisleri/ http://www.pearltrees.com/sporbahisleri https://pharmahub.org/members/26874/blog https://www.zupyak.com/u/Spor-Bahisleri/ https://www.metroflog.co/sporbahisleri https://www.fuzia.com/fz/spor-bahisleri https://tr.pinterest.com/sporbahislerim/ https://my.getjealous.com/sporbahisleri https://sporbahisleri.contently.com https://hubpages.com/@sporbahisleri https://www.tumblr.com/sporbahisleri https://hub.docker.com/u/sporbahislerim https://betsiteleri.blogfree.net https://betsiteleri.amebaownd.com https://sporbahisleri.pixnet.net/blog https://betsiteleri.seesaa.net https://betsiteleri.threadless.com https://betsiteleri.neocities.org https://bahissiteleri.localinfo.jp https://betsiteleri.shopinfo.jp https://teletype.in/@betsiteleri https://ubl.xml.org/users/sporbahisleri https://betsiteleri.educatorpages.com https://betsiteleri.onlc.fr https://sporbahisleri.gumroad.com
Introduction
Drought is a worldwide problem constraining global crop production and food quality, particularly in the drylands. Global climate change is modifying patterns of rainfall and temperature variation, thereby posing serious threats to agricultural productivity (Macholdt and Honermeier, 2016). This sitation has serious implications for farmers. Pakistan lies in a semi-arid region, and therefore, most of the agricultural activities of the country depend on supplemental irrigation (Abbas et al., 2015). The export of numerous value added cotton-based textiles sustains the economy of Pakistan, contributing 7.1% to the agriculture sector and 1.5% to GDP (Government of Pakistan, 2015). Owing to the importance of cotton for the local textile industry and export, it is widely grown in the core and non-core areas of the country irrigated by canals (Bhatti et al., 2009). Development of quality fiber depends on availability of adequate water. Therefore, cotton production declines if the crop encounters drought during the reproductive stage (Saleem et al., 2016; Wang et al., 2016). Because drought is a major threat to cotton production, it requires more attention than other problems. In such a scenario, breeding crop varieties with improved drought tolerance has great economic importance (Umezawa et al., 2006), and genetic variation is the most important prerequisite for successful accomplishment of this goal (Ullah et al., 2008; Azhar et al., 2009).
The advent of biotechnology has accelerated the process of plant breeding by introducing novel techniques for creation and exploitation of genetic variation. Among various biotechnological tools, DNA markers are extensively employed to study genetic diversity among populations. These markers detect DNA polymorphism, and they are considered reliable for molecular characterization of germplasm. Molecular markers like Random Amplified Polymorphic DNA (RAPD), Amplified Fragment Length Polymorphism (AFLP), Restriction Fragment Length Polymorphism (RFLP), Simple Sequence Repeats (SSR), Inter Simple Sequence Repeats (ISSR) and Sequence-related amplified polymorphism (SRAP) have been extensively used in analyzing cotton genotypes (Preetha and Raveendren, 2008; Bardak and Bolek, 2012). The potential of this technology was evident in a number of applications, such as marker assisted selection (MAS), QTL mapping, DNA fingerprinting, and hybrid verification in Gossypium species (Asif et al., 2009; Islam et al., 2012).
Simple Sequence Repeats (SSR) or microsatellites are short, tandemly repeated motifs (DNA sequences) with widespread distribution throughout the genome. Hoshino et al. (2012) reported SSR markers to be highly polymorphic, co-dominant, easy to interpret and transferable between species. In addition, according to Zhang et al. (2012) these markers are cost-effective and easily detectable through polymerase chain reaction (PCR) than the other marker techniques. SSR markers remain conserved among closely related species and therefore extensively used in selection of plants possessing desirable traits, genetic diversity analysis, assessment of phylogenetic relationships, linkage analysis, and cultivar identification (Morgante et al., 2002; Zhang et al., 2003).
In the present study, 22 cotton genotypes were analyzed using SSR markers to explore genetic divergence, with the objective of identifying the genetic background of the cotton genotypes.
Materials and Methods
Plant material
The present study was conducted at the Department of Plant Breeding and Genetics, University of Agriculture, Faisalabad, Pakistan. In this investigation, 50 cotton genotypes were subjected to artificial drought stress, equivalent to 80% of the field capacity, in a greenhouse. Drought stress was imposed at the two true leaves stage and continued for four weeks. After four weeks, data were recorded on drought induced morpho-physiological responses, including root length, shoot length, relative water content, and water content in excised leaves (data not shown here). Out of the 50 cotton genotypes studied, 22 were selected based on drought tolerance in the traits mentioned above. These 22 cotton genotypes (Table 1) were grown in the greenhouse. Seeds of all 22 cotton genotypes were soaked overnight. Holes of 2.5 cm depth were prepared in earthen pots containing a mixture of sand, soil, and farmyard manure in equal proportions. During germination and growth, temperature in the greenhouse was maintained at 20°C during night and 30°C during daytime using water circulation and electric heaters. The plants were provided a photoperiod of 16 h, and relative humidity of 65 ± 2%. Young leaves from three-week-old seedlings were collected in plastic zipper bags and stored in a freezer at -70°C for further use.
| Sr. No. | Genotypes | Sr. No. | Genotypes |
|---|---|---|---|
| 1 | CIM-496 | 12 | FH-900 |
| 2 | 149F | 13 | CIM-240 |
| 3 | DPL-26 | 14 | CIM-70 |
| 4 | BOU-1724 | 15 | CIM-109 |
| 5 | B-557 | 16 | BH-36 |
| 6 | FH-87 | 17 | NIAB-111 |
| 7 | FH-1000 | 18 | MNH-552 |
| 8 | NF-801-2-37 | 19 | 124F |
| 9 | MNH-129 | 20 | 4F |
| 10 | H-499 | 21 | CIM-707 |
| 11 | CIM-446 | 22 | BH-118 |
Table 1. Names of 22 cotton genotypes used in genetic diversity analysis based on SSR markers.
DNA isolation and quantification
Genomic DNA extraction was performed by the Cetyl trimethylammonium bromide (CTAB) method (Murray and Thompson, 1980). The concentration of extracted DNA was determined by spectrophotometry at 260 and 280 nm using the NanoDrop ND-1000 Spectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA). Quality of DNA was checked by electrophoresis of DNA samples on 0.8% agarose gel. The DNA samples that produced a smear on the gel were rejected. Working dilutions of DNA for SSR analysis (30 ng/μL), were prepared by adding double distilled H2O to aliquots of the stock DNA samples.
Microsatellite/SSR analysis
Thirty SSR primers of the JESPR series (Reddy et al., 2001) were used to identify the genetic divergence among the 22 genotypes. The final volume of each PCR mixture was 20 μL containing 2 μL template DNA, 6.4 μL 2.5 mM dNTPs, 2 μL (NH4)2SO4 buffer, 2 μL 25 mM MgCl2, 2 μL SSR primers (1 μL each of the forward and reverse primers), 0.2 μL Taq DNA polymerase, and 5.8 μL double distilled H2O. SSR amplification was performed in a Peqlab gradient thermocycler (BIO RAD Laboratories, Hercules, CA, USA). The PCR consisted of initial DNA denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 1 min, and ended with a final extension of 72°C for 4 min. The PCR products were stored at -20°C. All the SSR (PCR) fragments were resolved on a freshly prepared 6% polyacrylamide gel [acrylamide:bisacrylamide (20:1) mixture] using a vertical gel electrophoresis system (PROTEAN II xi Cell, BIO RAD Laboratories, Hercules, California, USA), using 1X Tris/Borate/Ethylenediaminetetraacetic acid TBE as running buffer at 200 V. The 50 bp DNA ladder (Fermentas, Waltham, MA, USA) was used to estimate the molecular weights of the amplified PCR products. Finally, the gel was immersed in a staining solution (8 μL ethidium bromide dissolved in 100 mL of distilled water) for 20 min, and washed with double distilled water for 5-10 s. Photographs of the gel were taken using Molecular Imager® Chemidoc (BIO RAD Laboratories, Hercules, CA, USA).
SSR data analysis
All visible and unambiguously identified fragments were scored as present (1) and absent (0). Polymorphic Information Content (PIC) value of each SSR locus was also calculated using the equation developed by Anderson et al. (1993).
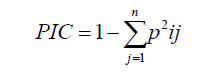 (Equation 1)
(Equation 1)
where pij is the frequency of jth allele for locus i. Mean band frequency (MBF) was computed by the following equation:
MBF = n / N (Equation 2)
where n is the number of plants carrying particular band; N is the total number of cultivars. Proportion of polymorphic bands was calculated as below:
PPB = Number of common bands / Total number of bands (Equation 3)
Popgene version 1.44 (Yeh and Boyle, 1997) was used to calculate the similarity indices using Nei’s similarity coefficient (Nei, 1972), and analyze the genetic relationship using cluster analysis.
Results and Discussion
Molecular marker technology holds great promise for crop improvement, particularly in cotton, a crop in which repeated cycles of directional selection for high yield and early fruiting might have caused loss of genetic diversity (Iqbal et al., 2001). In the present study, 30 SSR primer pairs were used to estimate genetic divergence among 22 cotton genotypes. Out of 30, 11 SSR primers (i.e., 37%) were informative and discriminated among the cotton genotypes (Table 2). These 11 polymorphic primers detected 41 alleles with an average of 3.72 alleles per locus. Among these 41 alleles, 16 (i.e., 39%) were polymorphic. PIC values ranged from 0.17-0.70. These findings were consistent with that of Bertini et al. (2006), who reported low PIC values while examining the genetic diversity in 53 cotton cultivars. In addition, Jia et al. (2014), who used 106 SSR primer pairs to estimate molecular diversity in 323 genotypes of Gossypium hirsutum grown under drought stress, detected narrow genetic base in the experimental material, indicated by low PIC value (0.53). Further, previous molecular markerbased studies had shown limited genetic variation in upland cotton. For instance, Rungis et al. (2005), Zhang et al. (2011), Ullah et al. (2012), and Dahab et al. (2013) assessed genetic distances among selected cotton cultivars using a limited number of microsatellites, and reported low allelic diversity. In contrast, studies conducted on cotton race stocks detected higher polymorphism, which clearly suggests that modern cultivars have narrow genetic base owing to domestication (Liu et al., 2000; Azmat and Khan, 2010).
| Sr No. | Primer name | Forward primer | Concen-tration of primer stocks (µg) | Repeat type | Number of alleles | PIC | MBF |
|---|---|---|---|---|---|---|---|
| 1 | JESPR247 F | 5'-GCTTCTTCCATTTTATTCAAG-3' | 328.7 | (CT)15 | 1 | 0.32 | 0.05 |
| JESPR247 R | 5'-CAGCGGCAACCAAAAAG-3' | 223.7 | |||||
| 2 | JESPR284 F | 5'-CAAGATCCATCTGCTGATTAG-3' | 285.1 | (CA)25(TA)5 | 1 | 0.50 | 0.05 |
| JESPR284 R | 5'-GTATATACAAGTATAAAGTATTGG-3' | 309.4 | |||||
| 3 | JESPR291 F | 5'-CATTCCCCACTTTGCTCTTAC-3' | 345.5 | (CTT)8 | 5 | 0.70 | 0.14 |
| JESPR291 R | 5'-CATGTTTCTTTGCCCATC-3' | 293.7 | |||||
| 4 | JESPR292 F | 5'-GCTTGCAATCTCCTACACC-3' | 297.4 | (CTT)7 | 1 | 0.50 | 0.05 |
| JESPR292 R | 5'-GAATATGTTTCATAGAATGGC-3' | 285.7 | |||||
| 5 | JESPR293 F | 5'-CGAGATTTTAAGATTGTGC-3' | 257.5 | (GAA)7 | 1 | 0.17 | 0.05 |
| JESPR293 R | 5'-TGATGGCAAAAGCACC-3' | 221.2 | |||||
| 6 | JESPR295 F | 5'-GCCTCGTTTAAGCCCATAAAC-3' | 289.0 | (CTT)7 | 1 | 0.33 | 0.05 |
| JESPR295 R | 5'-GAGGGCCATAGTCACCGG-3' | 229.9 | |||||
| 7 | JESPR300 F | 5'-CGCATCACAAACCAAACAC-3' | 259.5 | (CTT)5 | 1 | 0.20 | 0.05 |
| JESPR300 R | 5'-CGGAAAATGATGATGATGAAGAAG-3' | 320.6 | (CAT)6 | ||||
| 8 | JESPR301 F | 5'-TGAGTTCCGAATTCCTTGG-3' | 293.2 | (CAT)8 | 1 | 0.50 | 0.05 |
| JESPR301 R | 5'-CGGGCTAAGTGTTTTTCG-3' | 279.7 | |||||
| 9 | JESPR302 F | 5'-CACTCCTAGCTTCTTGGCATC-3' | 341.5 | (GAT)5 | 2 | 0.25 | 0.09 |
| JESPR302 R | 5'-CTGCGATCTTGGCACAG-3' | 245.9 | |||||
| 10 | JESPR304 F | 5'-GAAATGCATTCCCTCAAAAGC-3' | 285.6 | (GAT)8 | 1 | 0.33 | 0.05 |
| JESPR304 R | 5'-AGACTCTATCGAATGACCCTG-3' | 288.6 | |||||
| 11 | JESPR305 F | 5'-CGATCCATCAAAGGCGAC-3' | 268.1 | (GAT)6 | 1 | 0.50 | 0.05 |
| JESPR305 R | 5'-CCGCCTCAGCACCATTTAC-3' | 2847 |
Table 2. Details of 11 polymorphic SSR forward (F) and reverse (R) primers and their amplification products.
In the present study, genetic similarity coefficients ranged from 0.87-1.00. The cluster analysis (Figure 1) produced by un-weighted pair group method of arithmetic averages (UPGMA), grouped 21 genotypes in two main clusters, which further branched into subclusters. CIM-109 did not group with other genotypes and therefore exhibited maximum divergence. Clusters I and II contained eight and thirteen genotypes, respectively. In cluster I, NIAB-111 and BH-118 formed the most divergent pair, while DPL-26, CIM-496 and 124F were grouped together showing high level of genetic relatedness (similarity coefficient=1.00). In cluster II, maximum genetic relatedness was found in genotypes FH-1000, H-499, CIM- 446, FH-900, and CIM-240. In the same cluster, another group was constituted by genetically related genotypes including BOU-1724, FH-87, NF-801-2-37 and MNH-129 and CIM-707.
Figure 1: Cluster analysis showing the relationship among 22 cotton genotypes generated by 11 polymorphic SSR primers [1 = CIM-496, 2 =149F, 3 = DPL-26, 4 = BOU-1724, 5 = B-557, 6 = FH-87, 7 = FH-1000, 8 = NF-801-2- 37, 9 = MNH-129, 10 = H-499, 11 = CIM-446, 12 = FH-900, 13 = CIM-240, 14 = CIM-707, 15 = CIM-109, 16 = BH-36, 17 = NIAB-111, 18 = MNH-552, 19 = 124F, 20 = 4F, 21 = CIM-70, 22 = BH-118].
Similar findings were reported by Kalivas et al. (2011) and Ehsan et al. (2013) who detected low genetic diversity in commercial cotton cultivars used in their study.
The principal component analysis (PCA) plot, based on the first and second component factors of the microsatellite data, grouped the 22 cotton genotypes mainly into three groups (Figure 2). Group I was located near the X-axis, and contained genotypes BOU-1724, FH-87, NF-801-2-37 and MNH-129 and CIM-707. The distant location of group I from the origin indicated broad genetic base of genotypes included in it. However, the location of the genotypes B557 and CIM-707 close to the origin indicated lack of genetic diversity. The PCA plot positioned group II and group III near the Y-axis and the Y´-axis respectively. Genotypes included in group II [FH-1000, H-499, CIM-446, CIM-900, FH- 240] revealed more genetic divergence than those clustered in group III [MNH-552 and BH-118]. Thus, PCA plot confirmed the information generated by the genetic similarity matrix and cluster analysis.
Conclusion
In the present study, low genetic diversity was observed among all the cotton genotypes studied. The results of phenotypic screening did not validate SSR based selection, because drought tolerance specific primers were not used. However, the experimental material used in the present study was screened under drought stress conditions, which might be the cause of reduced genetic diversity.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
We are grateful for the financial support provided by Higher Education Commission, Islamabad, Pakistan for this research under HEC Indigenous Ph.D. Fellowship Program.
About the Authors
Corresponding Author
F.S. Awan
Centre for Agriculture Biochemistry & Biotechnolog, University of Agriculture, Faisalabad, Pakistan
- Email:
- awanfaisal@yahoo.com
References
- Abbas SH, Bhatti AA, Asif M, Islam Z, et al. (2015). Effect of supplemental irrigation on wheat water productivity under rainfed ecology of Pothohar, Pakistan. Innovare J. Agr. Sci. 3: 10-13.
- Anderson JA, Churchill GA, Autrique JE, Tanksley SD, et al. (1993). Optimizing parental selection for genetic linkage maps. Genome 36: 181-186. https://doi.org/10.1139/g93-024
- Asif M, Rahman M-U, Mirza JI and Zafar Y (2009). Parentage confirmation of cotton hybrids using molecular markers. Pak. J. Bot. 41: 695-701.
- Azhar FM, Ali Z, Akhtar M, Khan A, et al. (2009). Genetic variability of heat tolerance and its effect on yield and fibre quality traits in upland cotton (Gossypium hirsutum L.). Plant Breed. 128: 356-362. https://doi.org/10.1111/j.1439- 0523.2008.01574.x
- Azmat MA and Khan AA (2010). Assessment of genetic diversity among the varieties of Gossypium arboreum and Gossypium hirsutum through random amplification of polymorphic DNA (RAPD) and simple sequence repeat (SSR) markers. Pak. J. Bot. 42: 3173-3181.
- Bardak A and Bolek Y (2012). Genetic diversity of diploid and tetraploid cottons determined by SSR and ISSR markers. Turk. J. Field Crops 17: 139-144.
- Bertini CH, Schuster I, Sediyama T, Barros EGD, et al. (2006). Characterization and genetic diversity analysis of cotton cultivars using microsatellites. Genet. Mol. Biol. 29: 321-329. https://doi.org/10.1590/S1415-47572006000200021
- Bhatti AM, Suttinon P and Nasu S (2009). Agriculture water demand management in Pakistan: a review and perspective. J. Soc. Social Manage. Syst.: 1-7.
- Dahab AA, Saeed M, Mohamed BB, Ashraf MA, et al. (2013). Genetic diversity assessment of cotton (Gossypium hirsutum L.) genotypes from Pakistan using simple sequence repeat markers. Aust. J. Crop Sci. 7: 261-267.
- Ehsan B, Haque A, Younas M, Shaheen T, et al. (2013). Assessment of genomic diversity of cotton (Gossypium hirsutum) genotypes using simple sequence repeats markers through genetic analysis software. Int. J. Agric. Biol. 15: 968-972.
- Government of Pakistan (2015). Economic Affairs Wing, Ministry of Finance, Islamabad, Pakistan. Hoshino AA, Bravo JP, Nobile PM and Morelli KA (2012). Microsatellites as tools for genetic diversity analysis. In: Genetic diversity in microorganisms (Caliskan M eds.). In Tech, Croatia, 149-170.
- Iqbal MJ, Reddy OUK, El-Zik KM and Pepper AE (2001). A genetic bottleneck in the evolution under domestication of upland cotton Gossypium hirsutum L. examined using DNA fingerprinting. Theor. Appl. Genet. 103: 547-554. https:// doi.org/10.1007/PL00002908
- Islam MN, Molla M, Rohman M, Hasanuzzaman M, et al. (2012). DNA fingerprinting and genotyping of cotton varieties using SSR Markers. Not. Bot. Horti Agrobot. Cluj-Napoca 40: 261-265.
- Jia Y-H, Sun J-L, Wang X-W, Zhou Z-Li, et al. (2014). Molecular diversity and association analysis of drought and salt tolerance in Gossypium hirsutum L. germplasm. J. Integr. Agric. 13: 1845-185. https://doi.org/10.1016/S2095- 3119(13)60668-1
- Kalivas A, Xanthopoulos F, Kehagia O and Tsaftaris AS (2011). Agronomic characterization, genetic diversity and association analysis of cotton cultivars using simple sequence repeat molecular markers. Genet. Mol. Res. 10: 208- 217. https://doi.org/10.4238/vol10-1gmr998
- Liu S, Cantrell RG, McCarty JC and Stewart JM (2000). Simple sequence repeat based assessment of genetic diversity in cotton race stock accessions. Crop Sci. 40: 1459-1469. https://doi.org/10.2135/cropsci2000.4051459x
- Macholdt J and Honermeier B (2016). Impact of climate change on cultivar choice: adaptation strategies of farmers and advisors in German cereal production. Agronomy (Basel) 6: 1-40. https://doi.org/10.3390/agronomy6030040
- Morgante M, Hanafey M and Powell W (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30: 194-200. https://doi.org/10.1038/ng822
- Murray MG and Thompson WF (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321- 4325. https://doi.org/10.1093/nar/8.19.4321
- Nei M (1972). Genetic distance between populations. Am. Nat. •••: 283-292. https://doi.org/10.1086/282771 Preetha S and Raveendren TS (2008). Molecular marker technology in cotton. Biotech. Mol. Biol. Rev. 3: 32-45.
- Reddy OUK, Pepper AE, Abdurakhmonov IY, Saha S, et al. (2001). The identification of dinucleotide and trinucleotide microsatellite repeat loci from cotton G. hirsutum L. J. Cotton Sci. 5: 103-113.
- Rungis D, Llewellyn D, Dennis ES and Lyon BR (2005). Simple sequence repeat (SSR) markers reveal low levels of polymorphism between cotton (Gossypium hirsutum L.) cultivars. Crop Pasture Sci. 56: 301-307. https://doi. org/10.1071/AR04190
- Saleem MF, Raza MAS, Ahmad S, Khan IH, et al. (2016). Understanding and mitigating the impacts of drought stress in cotton- a review. Pak. J. Agric. Sci. 53: 609-623.
- Ullah I, Ashraf M and Zafar Y (2008). Genotypic variation for drought tolerance in cotton (Gossypium hirsutum L.): Leaf gas exchange and productivity. Flora-Morph. Dist. Func. Ecol. Plants 203: 105-115. https://doi.org/10.1016/j. flora.2007.12.001
- Ullah I, Iram A, Iqbal MZ, Nawaz M, et al. (2012). Genetic diversity analysis of Bt cotton genotypes in Pakistan using simple sequence repeat markers. Genet. Mol. Res. 11: 597-605. https://doi.org/10.4238/2012.March.14.3
- Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, et al. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17: 113-122. https://doi.org/10.1016/j. copbio.2006.02.002
- Wang R, Ji S, Zhang P, Meng Y, et al. (2016). Drought effects on cotton yield and fiber quality on different fruiting branches. Crop Sci. 56: 1265-1276. https://doi.org/10.2135/cropsci2015.08.0477
- Yeh FC and Boyle TGB (1997). Population genetic analysis of co-dominant and dominant markers and quantitative traits. J. Bot. 129: 157-163.
- Zhang Q, Li J, Zhao Y, Korban SS, et al. (2012). Evaluation of genetic diversity in Chinese wild apple species along with apple cultivars using SSR markers. Plant Mol. Biol. Report. 30: 539-546. https://doi.org/10.1007/s11105-011-0366-6
- Zhang T, Yuan Y, Yu J, Guo W, et al. (2003). Molecular tagging of a major QTL for fiber strength in Upland cotton and its marker-assisted selection. Theor. Appl. Genet. 106: 262-268. https://doi.org/10.1007/s00122-002-1101-3
- Zhang Y, Wang XF, Li ZK, Zhang GY, et al. (2011). Assessing genetic diversity of cotton cultivars using genomic and newly developed expressed sequence tag-derived microsatellite markers. Genet. Mol. Res. 10: 1462-1470. https:// doi.org/10.4238/vol10-3gmr1277
Keywords:
Download:
Full PDF- Share This

